Hmga2 is necessary for Otx2-dependent exit of embryonic stem cells from the pluripotent ground state
- PMID: 27036552
- PMCID: PMC4818510
- DOI: 10.1186/s12915-016-0246-5
Hmga2 is necessary for Otx2-dependent exit of embryonic stem cells from the pluripotent ground state
Abstract
Background: A crucial event in the differentiation of mouse embryonic stem cells (ESCs) is the exit from the pluripotent ground state that leads to the acquisition of the 'primed' pluripotent phenotype, characteristic of the epiblast-like stem cells (EpiLCs). The transcription factors Oct4 and Otx2 play a key role in this phenomenon. In particular, Otx2 pioneers and activates new enhancers, which are silent in ESCs and which control the transcription of genes responsible for the acquisition of the EpiLC phenotype. An important point that remains to be addressed is the mechanism through which Otx2 engages the new enhancers and stably associates with them. Hmga2 is a member of the high-mobility group family of proteins, non-histone components of chromatin whose expression is high during embryogenesis and becomes low or undetectable in adults. Its high expression during embryogenesis suggests that Hmga2 fulfills important roles in development.
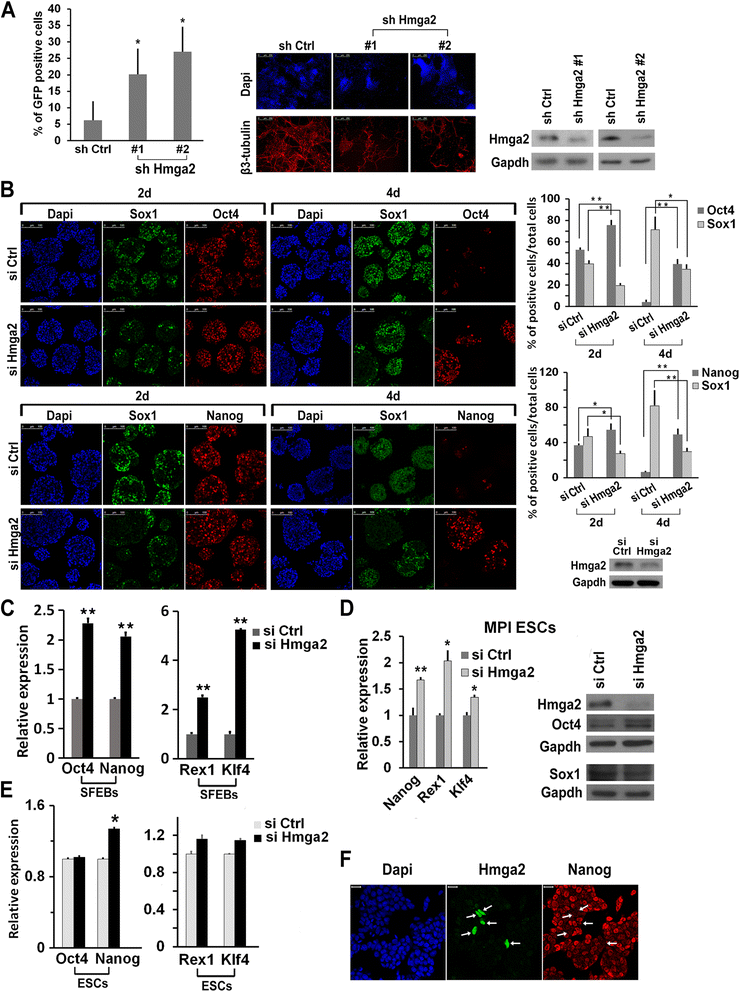
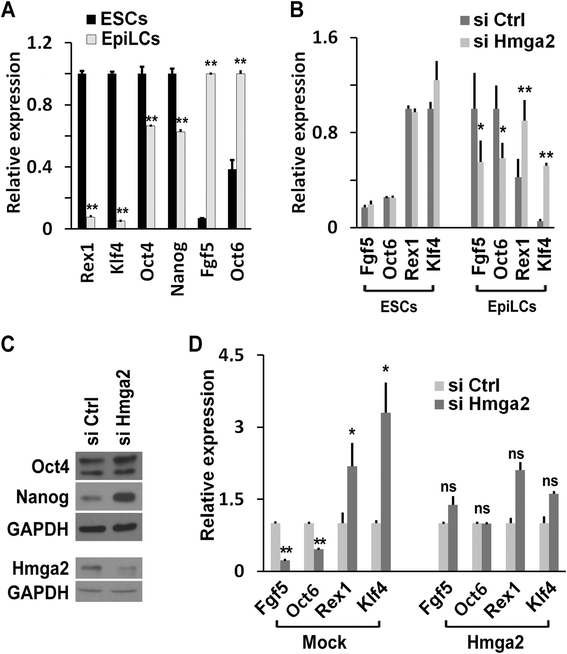
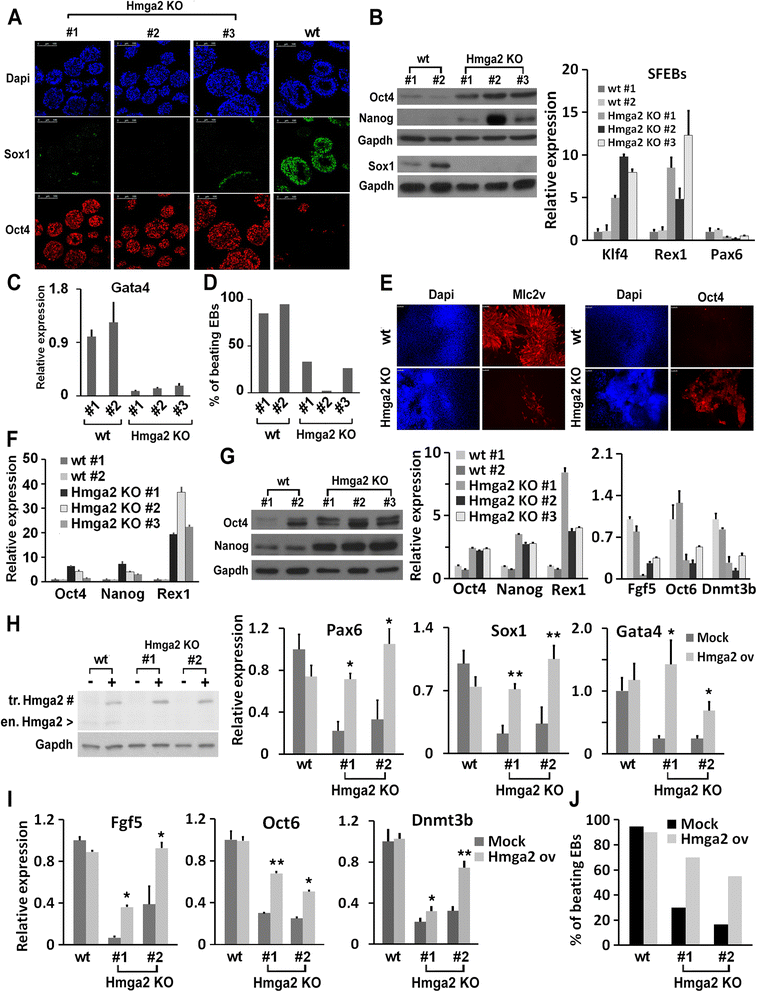
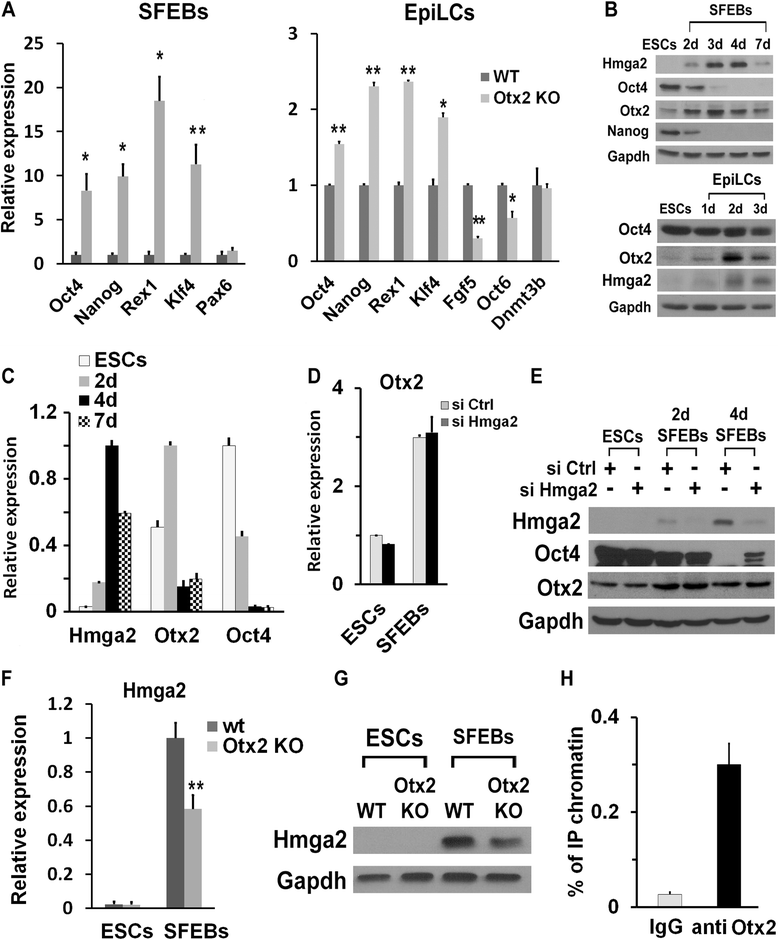
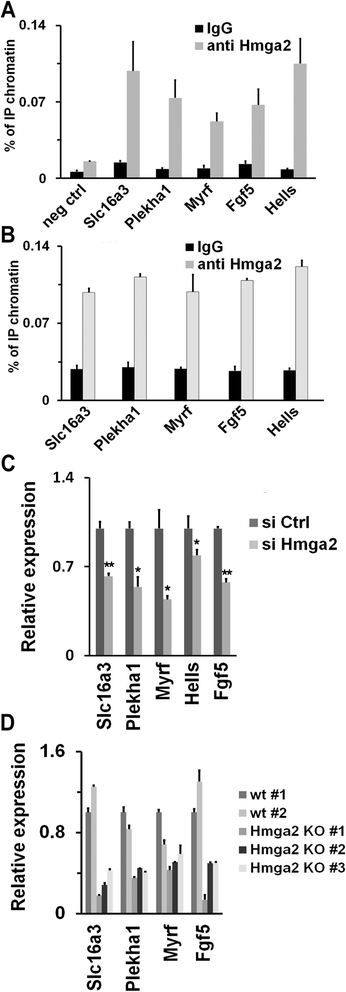
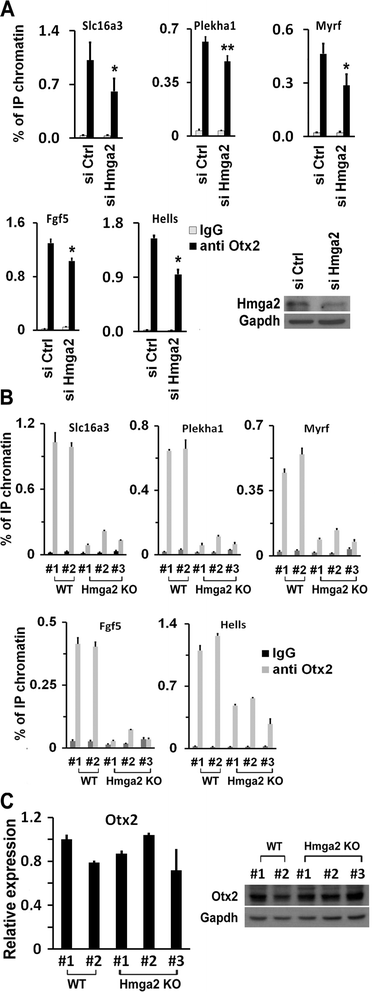
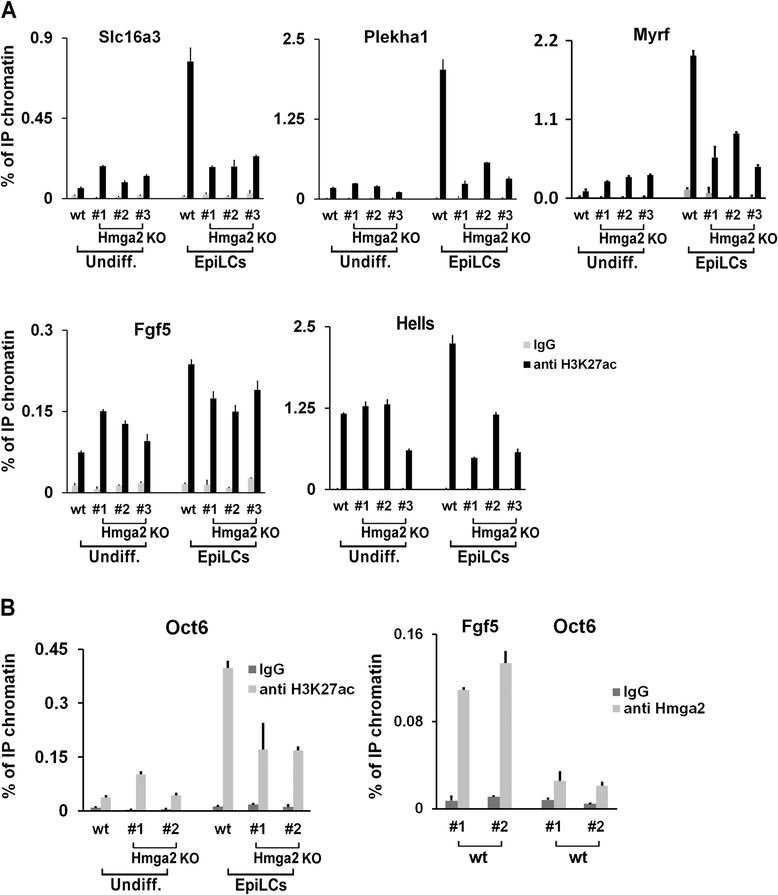
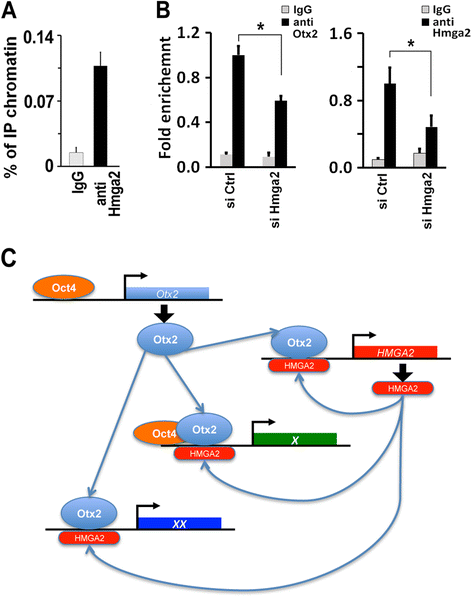
Results: Here, we demonstrate that Hmga2 accumulates soon after the induction of ESC differentiation. Its suppression hampers the exit of ESCs from the pluripotent ground state and their differentiation into EpiLCs. Mechanistically, Hmga2 controls the differentiation process by cooperating with Otx2 in the pioneering of new enhancers. In Hmga2 null induced pluripotent stem cells we observe that Otx2 fails to regulate its target genes upon the induction of differentiation. Hmga2 associates to Otx2-bound loci in EpiLCs, and in Hmga2 KO cells Otx2 is unable to engage and activate the new enhancers, thus indicating that Hmga2 is required for the binding of Otx2 to its cis-elements. We find that this mechanism also operates on the Hmga2 gene, which is one of the targets of Otx2, thus indicating the existence of a positive feedback loop.
Conclusions: Our findings reveal a novel mechanism necessary for the exit of ESCs from the pluripotent ground state. Upon the induction of ESC differentiation, Otx2 alone or in combination with Oct4 engages new enhancers, which are silent in undifferentiated ESCs. The Hmga2 gene is activated by Otx2 and Hmga2 protein binds to the enhancers targeted by Otx2, thus facilitating the engagement and/or the stable association of Otx2. Therefore, our results demonstrate that Hmga2 is a key element of the regulatory network that governs the exit of ESCs from the pluripotent ground state.
Keywords: Embryonic stem cell differentiation; Enhancer activation; High-mobility group protein.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

