The microbiome of urban waters
- PMID: 27036741
- PMCID: PMC8793681
- DOI: 10.2436/20.1501.01.244
The microbiome of urban waters
Abstract
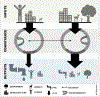
More than 50% of the world's population lives in urban centers. As collection basins for landscape activity, urban waters are an interface between human activity and the natural environment. The microbiome of urban waters could provide insight into the impacts of pollution, the presence of human health risks, or the potential for long-term consequences for these ecosystems and the people who depend upon them. An integral part of the urban water cycle is sewer infrastructure. Thousands of miles of pipes line cities as part of wastewater and stormwater systems. As stormwater and sewage are released into natural waterways, traces of human and animal microbiomes reflect the sources and magnitude of fecal pollution and indicate the presence of pollutants, such as nutrients, pathogens, and chemicals. Non-fecal organisms are also released as part of these systems. Runoff from impervious surfaces delivers microbes from soils, plants and the built environment to stormwater systems. Further, urban sewer infrastructure contains its own unique microbial community seemingly adapted to this relatively new artificial habitat. High microbial densities are conveyed via pipes to waterways, and these organisms can be found as an urban microbial signature imprinted on the natural community of rivers and urban coastal waters. The potential consequences of mass releases of non-indigenous microorganisms into natural waters include creation of reservoirs for emerging human pathogens, altered nutrient flows into aquatic food webs, and increased genetic exchange between two distinct gene pools. This review highlights the recent characterization of the microbiome of urban sewer and stormwater infrastructure and its connection to and potential impact upon freshwater systems.
Keywords: aquatic food webs; human health; infrastructure and sanitation; next generation sequencing; urban freshwaters.
Copyright© by the Spanish Society for Microbiology and Institute for Catalan Studies.
Conflict of interest statement
Figures

References
-
- Cai L, Ju F, Zhang T (2014) Tracking human sewage microbiome in a municipal wastewater treatment plant. Appl Microbiol Biot 98:3317–3326 - PubMed
-
- Cao Y, Van De Werfhorst LC, Sercu B, Murray JL, Holden PA (2011) Application of an integrated community analysis approach for microbial source tracking in a coastal creek. Environ Sci Technol 45:7195–7201 - PubMed
-
- Cao Y, Van De Werfhorst LC, Dubinsky EA, Badgley BD, Sadowsky MJ, Andersen GL, Griffith JF, Holden PA (2013) Evaluation of molecular community analysis methods for discerning fecal sources and human waste. Water Res 47:6862–6872 - PubMed
-
- Caplin JL, Hanlon GW, Taylor HD (2008) Presence of vancomycin and ampicillin-resistant Enterococcus faecium of epidemic clonal complex-17 in wastewaters from the south coast of England. Environ Microbiol 10:885–892 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

