Proton irradiation induces persistent and tissue-specific DNA methylation changes in the left ventricle and hippocampus
- PMID: 27036964
- PMCID: PMC4815246
- DOI: 10.1186/s12864-016-2581-x
Proton irradiation induces persistent and tissue-specific DNA methylation changes in the left ventricle and hippocampus
Abstract
Background: Proton irradiation poses a potential hazard to astronauts during and following a mission, with post-mitotic cells at most risk because they cannot dilute resultant epigenetic changes via cell division. Persistent epigenetic changes that result from environmental exposures include gains or losses of DNA methylation of cytosine, which can impact gene expression. In the present study, we compared the long-term epigenetic effects of whole body proton irradiation in the mouse hippocampus and left ventricle. We used an unbiased genome-wide DNA methylation study, involving ChIP-seq with antibodies to 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) to identify DNA regions in which methylation levels have changed 22 weeks after a single exposure to proton irradiation. We used DIP-Seq to profile changes in genome-wide DNA methylation and hydroxymethylation following proton irradiation. In addition, we used published RNAseq data to assess whether differentially methylated regions were linked to changes in gene expression.
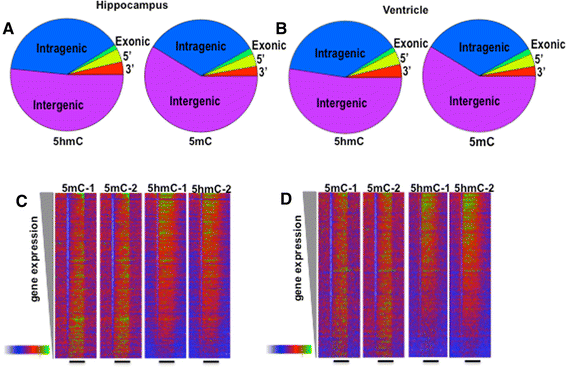
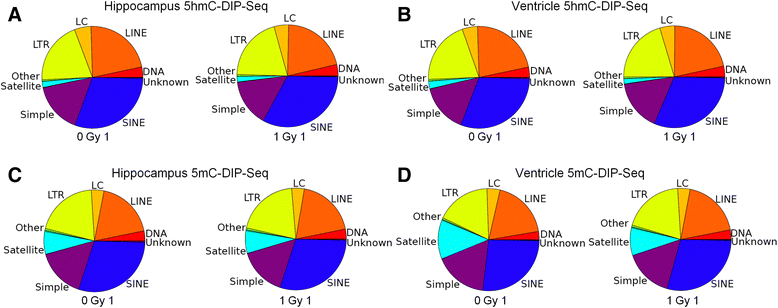
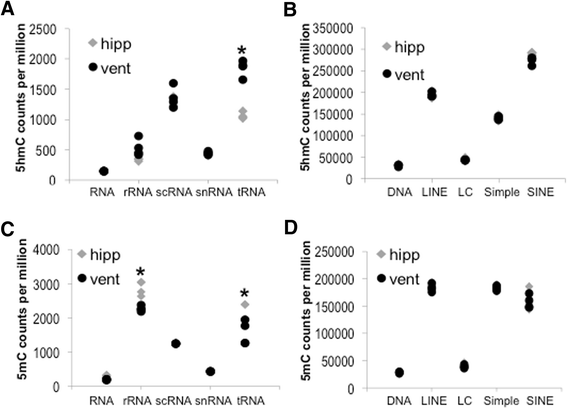
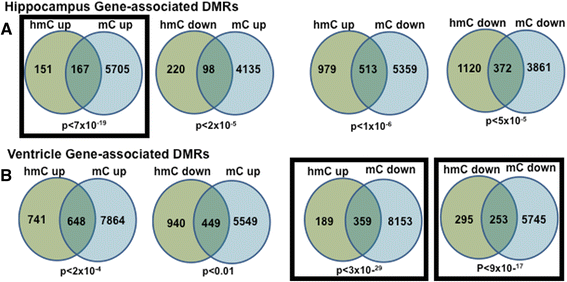
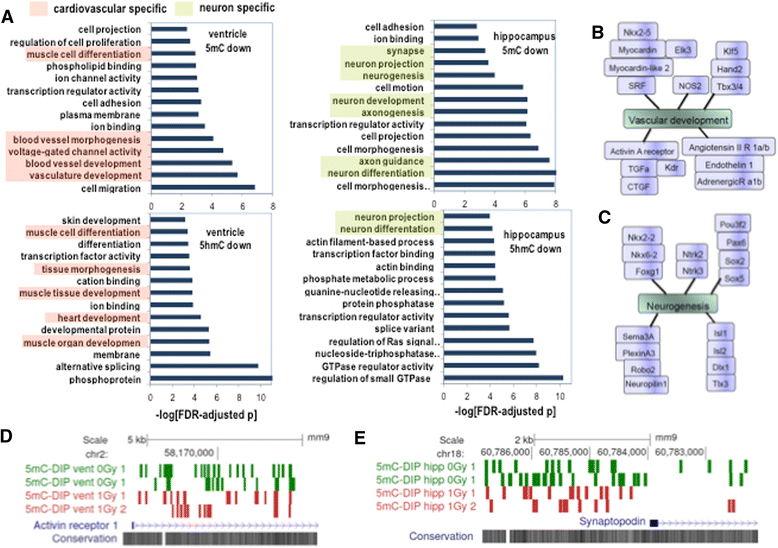
Results: The DNA methylation data showed tissue-dependent effects of proton irradiation and revealed significant major pathway changes in response to irradiation that are related to known pathophysiologic processes. Many regions affected in the ventricle mapped to genes involved in cardiovascular function pathways, whereas many regions affected in the hippocampus mapped to genes involved in neuronal functions. In the ventricle, increases in 5hmC were associated with decreases in 5mC. We also observed spatial overlap for regions where both epigenetic marks decreased in the ventricle. In hippocampus, increases in 5hmC were most significantly correlated (spatially) with regions that had increased 5mC, suggesting that deposition of hippocampal 5mC and 5hmC may be mechanistically coupled.
Conclusions: The results demonstrate long-term changes in DNA methylation patterns following a single proton irradiation, that these changes are tissue specific, and that they map to pathways consistent with tissue specific responses to proton irradiation. Further, the results suggest novel relationships between changes in 5mC and 5hmC.
Keywords: DNA methylation; Epigenetic; Hippocampus; Left ventricle; Proton irradiation; RNAseq; Ventricle.
Figures
References
-
- Szyf M. Epigenetics, DNA Methylation, and Chromatin Modifying Drugs. Annu Rev Pharmacol Toxicol. 2008. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

