Using ClinVar as a Resource to Support Variant Interpretation
- PMID: 27037489
- PMCID: PMC4832236
- DOI: 10.1002/0471142905.hg0816s89
Using ClinVar as a Resource to Support Variant Interpretation
Abstract
ClinVar is a freely accessible, public archive of reports of the relationships among genomic variants and phenotypes. To facilitate evaluation of the clinical significance of each variant, ClinVar aggregates submissions of the same variant, displays supporting data from each submission, and determines if the submitted clinical interpretations are conflicting or concordant. The unit describes how to (1) identify sequence and structural variants of interest in ClinVar by multiple searching approaches, including Variation Viewer and (2) understand the display of submissions to ClinVar and the evidence supporting each interpretation. By following this protocol, ClinVar users will be able to learn how to incorporate the wealth of resources and knowledge in ClinVar into variant curation and interpretation.
Keywords: ClinVar; clinical genetics; data sharing; databases; variant interpretation.
Copyright © 2016 John Wiley & Sons, Inc.
Figures








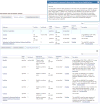
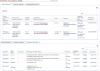
References
-
- den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–124. - PubMed
-
- Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13:680–685. - PubMed
-
- Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, Farrell CM, Hart J, Landrum MJ, McGarvey KM, Murphy MR, O'Leary NA, Pujar S, Rajput B, Rangwala SH, Riddick LD, Shkeda A, Sun H, Tamez P, Tully RE, Wallin C, Webb D, Weber J, Wu W, DiCuccio M, Kitts P, Maglott DR, Murphy TD, Ostell JM. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–763. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

