IL-8 induces miR-424-5p expression and modulates SOCS2/STAT5 signaling pathway in oral squamous cell carcinoma
- PMID: 27038552
- PMCID: PMC5423170
- DOI: 10.1016/j.molonc.2016.03.001
IL-8 induces miR-424-5p expression and modulates SOCS2/STAT5 signaling pathway in oral squamous cell carcinoma
Abstract
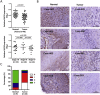
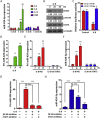
Suppressor of cytokine signaling (SOCS) proteins are negative feedback regulators of the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway. Dysregulation of SOCS protein expression in cancers can be one of the mechanisms that maintain STAT activation, but this mechanism is still poorly understood in oral squamous cell carcinoma (OSCC). Here, we report that SOCS2 protein is significantly downregulated in OSCC patients and its levels are inversely correlated with miR-424-5p expression. We identified the SOCS2 protein, which modulates STAT5 activity, as a direct target of miR-424-5p. The miR-424-5p-induced STAT5 phosphorylation, matrix metalloproteinases (MMPs) expression, and cell migration and invasion were blocked by SOCS2 restoration, suggesting that miR-424-5p exhibits its oncogenic activity through negatively regulating SOCS2 levels. Furthermore, miR-424-5p expression could be induced by the cytokine IL-8 primarily through enhancing STAT5 transcriptional activity rather than NF-κB signaling. Antagomir-mediated inactivation of miR-424-5p prevented the IL-8-induced cell migration and invasion, indicating that miR-424-5p is required for IL-8-induced cellular invasiveness. Taken together, these data indicate that STAT5-dependent expression of miR-424-5p plays an important role in mediating IL-8/STAT5/SOCS2 feedback loop, and scavenging miR-424-5p function using antagomir may have therapeutic potential for the treatment of OSCC.
Keywords: IL-8; Invasion; Oral squamous cell carcinoma; SOCS2; microRNA.
Copyright © 2016 The Authors. Published by Elsevier B.V. All rights reserved.
Figures




References
-
- Agrawal, N. , Frederick, M.J. , Pickering, C.R. , Bettegowda, C. , Chang, K. , Li, R.J. , Fakhry, C. , Xie, T.X. , Zhang, J. , Wang, J. , Zhang, N. , El-Naggar, A.K. , Jasser, S.A. , Weinstein, J.N. , Trevino, L. , Drummond, J.A. , Muzny, D.M. , Wu, Y. , Wood, L.D. , Hruban, R.H. , Westra, W.H. , Koch, W.M. , Califano, J.A. , Gibbs, R.A. , Sidransky, D. , Vogelstein, B. , Velculescu, V.E. , Papadopoulos, N. , Wheeler, D.A. , Kinzler, K.W. , Myers, J.N. , 2011. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 333, 1154–1157. - PMC - PubMed
-
- Ahomadegbe, J.C. , Barrois, M. , Fogel, S. , Le Bihan, M.L. , Douc-Rasy, S. , Duvillard, P. , Armand, J.P. , Riou, G. , 1995. High incidence of p53 alterations (mutation, deletion, overexpression) in head and neck primary tumors and metastases; absence of correlation with clinical outcome. Frequent protein overexpression in normal epithelium and in early non-invasive lesions. Oncogene. 10, 1217–1227. - PubMed
-
- Benekli, M. , Baer, M.R. , Baumann, H. , Wetzler, M. , 2003. Signal transducer and activator of transcription proteins in leukemias. Blood. 101, 2940–2954. - PubMed
-
- Britschgi, A. , Andraos, R. , Brinkhaus, H. , Klebba, I. , Romanet, V. , Muller, U. , Murakami, M. , Radimerski, T. , Bentires-Alj, M. , 2012. JAK2/STAT5 inhibition circumvents resistance to PI3K/mTOR blockade: a rationale for cotargeting these pathways in metastatic breast cancer. Cancer cell. 22, 796–811. - PubMed
-
- Calin, G.A. , Croce, C.M. , 2006. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 66, 7390–7394. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

