Reproducibility of Illumina platform deep sequencing errors allows accurate determination of DNA barcodes in cells
- PMID: 27038897
- PMCID: PMC4818877
- DOI: 10.1186/s12859-016-0999-4
Reproducibility of Illumina platform deep sequencing errors allows accurate determination of DNA barcodes in cells
Abstract
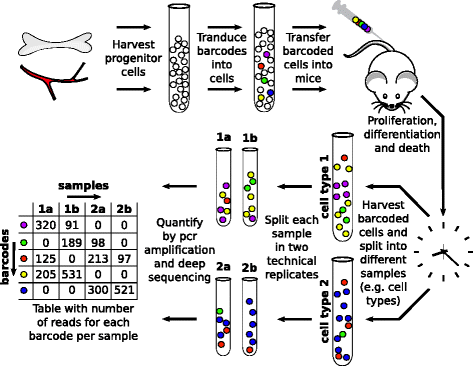
Background: Next generation sequencing (NGS) of amplified DNA is a powerful tool to describe genetic heterogeneity within cell populations that can both be used to investigate the clonal structure of cell populations and to perform genetic lineage tracing. For applications in which both abundant and rare sequences are biologically relevant, the relatively high error rate of NGS techniques complicates data analysis, as it is difficult to distinguish rare true sequences from spurious sequences that are generated by PCR or sequencing errors. This issue, for instance, applies to cellular barcoding strategies that aim to follow the amount and type of offspring of single cells, by supplying these with unique heritable DNA tags.
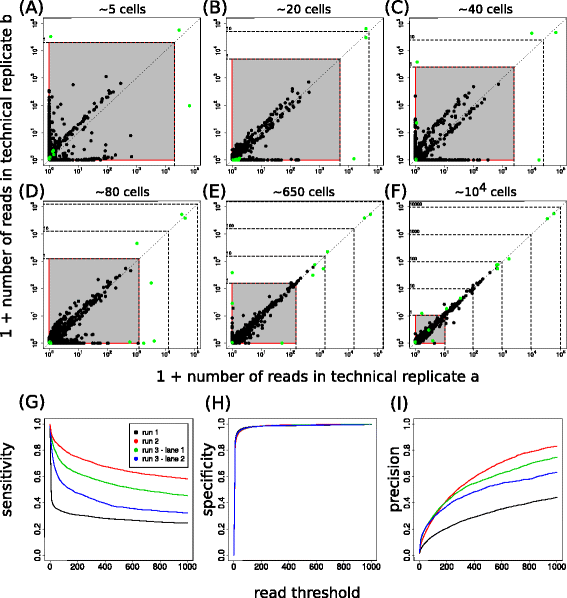
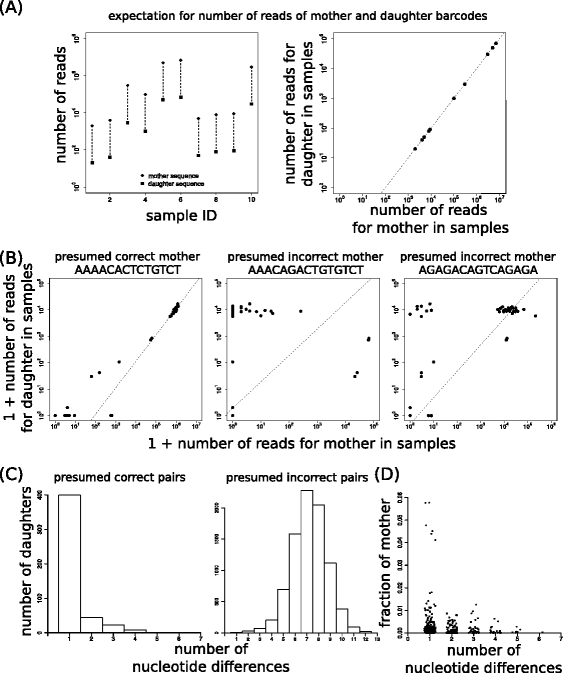
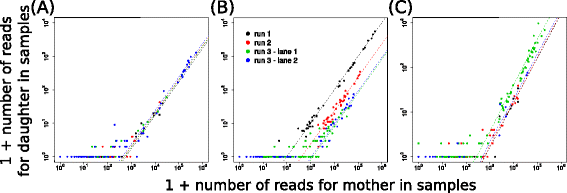
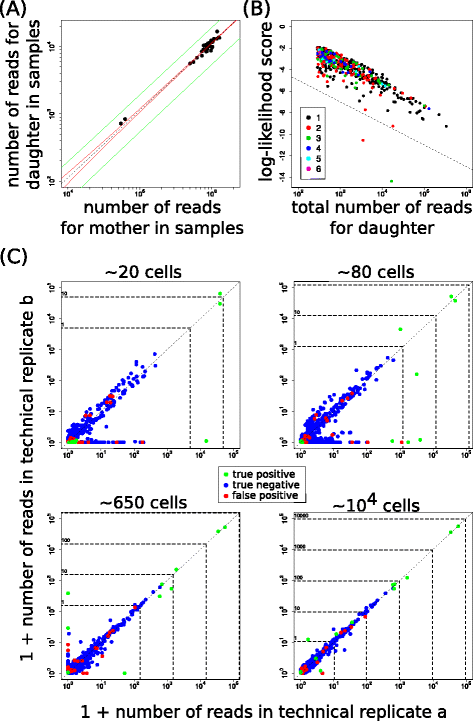
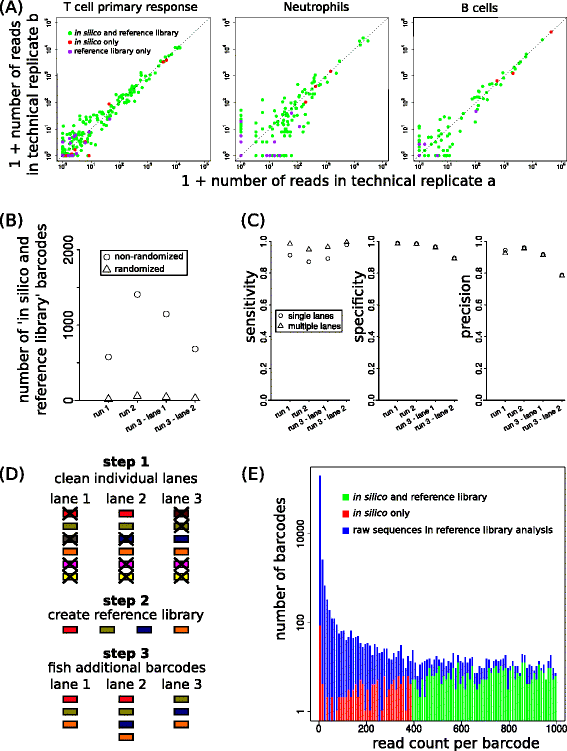
Results: Here, we use genetic barcoding data from the Illumina HiSeq platform to show that straightforward read threshold-based filtering of data is typically insufficient to filter out spurious barcodes. Importantly, we demonstrate that specific sequencing errors occur at an approximately constant rate across different samples that are sequenced in parallel. We exploit this observation by developing a novel approach to filter out spurious sequences.
Conclusions: Application of our new method demonstrates its value in the identification of true sequences amongst spurious sequences in biological data sets.
Keywords: Cellular barcoding; Illumina; Lineage tracing; Next generation sequencing; PCR error; Sequencing error.
Figures
Similar articles
-
Primer ID Validates Template Sampling Depth and Greatly Reduces the Error Rate of Next-Generation Sequencing of HIV-1 Genomic RNA Populations.J Virol. 2015 Aug;89(16):8540-55. doi: 10.1128/JVI.00522-15. Epub 2015 Jun 3. J Virol. 2015. PMID: 26041299 Free PMC article.
-
Barcode-free next-generation sequencing error validation for ultra-rare variant detection.Nat Commun. 2019 Feb 28;10(1):977. doi: 10.1038/s41467-019-08941-4. Nat Commun. 2019. PMID: 30816127 Free PMC article.
-
Illumina midi-barcodes: quality proof and applications.Mitochondrial DNA A DNA Mapp Seq Anal. 2019 Apr;30(3):490-499. doi: 10.1080/24701394.2018.1551386. Epub 2019 Jan 11. Mitochondrial DNA A DNA Mapp Seq Anal. 2019. PMID: 30633607
-
Cellular barcoding: lineage tracing, screening and beyond.Nat Methods. 2018 Nov;15(11):871-879. doi: 10.1038/s41592-018-0185-x. Epub 2018 Oct 30. Nat Methods. 2018. PMID: 30377352 Review.
-
Beyond genome sequencing: lineage tracking with barcodes to study the dynamics of evolution, infection, and cancer.Genomics. 2014 Dec;104(6 Pt A):417-30. doi: 10.1016/j.ygeno.2014.09.005. Epub 2014 Sep 28. Genomics. 2014. PMID: 25260907 Review.
Cited by
-
Clonal barcoding with qPCR detection enables live cell functional analyses for cancer research.Nat Commun. 2022 Jul 4;13(1):3837. doi: 10.1038/s41467-022-31536-5. Nat Commun. 2022. PMID: 35788590 Free PMC article.
-
Extracting, filtering and simulating cellular barcodes using CellBarcode tools.Nat Comput Sci. 2024 Feb;4(2):128-143. doi: 10.1038/s43588-024-00595-7. Epub 2024 Feb 19. Nat Comput Sci. 2024. PMID: 38374363 Free PMC article.
-
Systematic evaluation of error rates and causes in short samples in next-generation sequencing.Sci Rep. 2018 Jul 19;8(1):10950. doi: 10.1038/s41598-018-29325-6. Sci Rep. 2018. PMID: 30026539 Free PMC article.
-
A committed tissue-resident memory T cell precursor within the circulating CD8+ effector T cell pool.J Exp Med. 2020 Oct 5;217(10):e20191711. doi: 10.1084/jem.20191711. J Exp Med. 2020. PMID: 32728699 Free PMC article.
-
Limitations and challenges of genetic barcode quantification.Sci Rep. 2017 Mar 3;7:43249. doi: 10.1038/srep43249. Sci Rep. 2017. PMID: 28256524 Free PMC article.
References
-
- Brady T, Roth SL, Malani N, Wang GP, Berry CC, Leboulch P, Hacein-Bey-Abina S, Cavazzana-Calvo M, Papapetrou EP, Sadelain M, Savilahti H, Bushman FD. A method to sequence and quantify DNA integration for monitoring outcome in gene therapy. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkr140. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

