CD300c is uniquely expressed on CD56 bright Natural Killer Cells and differs from CD300a upon ligand recognition
- PMID: 27040328
- PMCID: PMC4819222
- DOI: 10.1038/srep23942
CD300c is uniquely expressed on CD56 bright Natural Killer Cells and differs from CD300a upon ligand recognition
Abstract
Paired receptors on NK cells recognize similar ligands with varied strength of binding ability and perform different functions. The CD300 molecules are emerging as novel immune regulators in health and disease due to their interaction with their lipid-nature ligands. Particularly, the paired receptors CD300c and CD300a have been shown to elicit activating and inhibitory capabilities, respectively. In the current study, we seek to investigate the expression and function of CD300c on human NK cells. We demonstrate that IL-2 and IL-15 treatment significantly induce CD300c expression exclusively on CD56(bright) NK cells. CD300c up-regulation requires STAT5 and its expression is inhibited by IL-4. Consistently, IL-2 secreted from activated CD4(+) T cells specifically induces the expression of CD300c on CD56(bright) NK cells. Crosslinking CD300c with a specific antibody enhances the proficiency of CD56(bright) NK cells to degranulate and induce chemokine and cytokine secretion. We also show the differential binding of CD300a and CD300c to their ligands phosphatidylethanolamine (PE) and phosphatidylserine (PS) and their differential ability to affect CD56(bright) NK cell functions. Our results provide an insight into the novel set of paired receptors CD300a and CD300c that are distinctively expressed on CD56(bright) NK cells with varied effector functions.
Figures

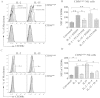


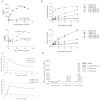

Similar articles
-
The interaction between CD300a and phosphatidylserine inhibits tumor cell killing by NK cells.Eur J Immunol. 2013 Aug;43(8):2151-61. doi: 10.1002/eji.201343433. Epub 2013 Jun 21. Eur J Immunol. 2013. PMID: 23640773 Free PMC article.
-
Human CD300C delivers an Fc receptor-γ-dependent activating signal in mast cells and monocytes and differs from CD300A in ligand recognition.J Biol Chem. 2013 Mar 15;288(11):7662-7675. doi: 10.1074/jbc.M112.434746. Epub 2013 Jan 31. J Biol Chem. 2013. PMID: 23372157 Free PMC article.
-
Interleukin-21 differentially affects human natural killer cell subsets.Immunology. 2007 Dec;122(4):486-95. doi: 10.1111/j.1365-2567.2007.02675.x. Epub 2007 Jul 16. Immunology. 2007. PMID: 17635612 Free PMC article.
-
The Biology and Disease Relevance of CD300a, an Inhibitory Receptor for Phosphatidylserine and Phosphatidylethanolamine.J Immunol. 2015 Jun 1;194(11):5053-60. doi: 10.4049/jimmunol.1500304. J Immunol. 2015. PMID: 25980030 Review.
-
Role of chemokines in the biology of natural killer cells.Curr Top Microbiol Immunol. 2010;341:37-58. doi: 10.1007/82_2010_20. Curr Top Microbiol Immunol. 2010. PMID: 20369317 Review.
Cited by
-
Altered Expression of CD300a Inhibitory Receptor on CD4+ T Cells From Human Immunodeficiency Virus-1-Infected Patients: Association With Disease Progression Markers.Front Immunol. 2018 Jul 23;9:1709. doi: 10.3389/fimmu.2018.01709. eCollection 2018. Front Immunol. 2018. PMID: 30083165 Free PMC article.
-
Genetic Correlation Analysis and Transcriptome-wide Association Study Suggest the Overlapped Genetic Mechanism between Gout and Attention-deficit Hyperactivity Disorder.Can J Psychiatry. 2021 Dec;66(12):1077-1084. doi: 10.1177/0706743720970844. Epub 2020 Nov 6. Can J Psychiatry. 2021. PMID: 33155823 Free PMC article.
-
The expression and function of human CD300 receptors on blood circulating mononuclear cells are distinct in neonates and adults.Sci Rep. 2016 Sep 6;6:32693. doi: 10.1038/srep32693. Sci Rep. 2016. PMID: 27595670 Free PMC article.
-
The role of phosphatidylserine on the membrane in immunity and blood coagulation.Biomark Res. 2022 Jan 15;10(1):4. doi: 10.1186/s40364-021-00346-0. Biomark Res. 2022. PMID: 35033201 Free PMC article. Review.
-
Inhibitory Receptors and Immune Checkpoints Regulating Natural Killer Cell Responses to Cancer.Cancers (Basel). 2021 Aug 24;13(17):4263. doi: 10.3390/cancers13174263. Cancers (Basel). 2021. PMID: 34503073 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

