Control of the collective migration of enteric neural crest cells by the Complement anaphylatoxin C3a and N-cadherin
- PMID: 27041467
- PMCID: PMC4937886
- DOI: 10.1016/j.ydbio.2016.03.022
Control of the collective migration of enteric neural crest cells by the Complement anaphylatoxin C3a and N-cadherin
Abstract
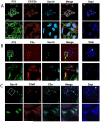
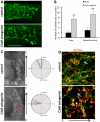
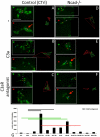
We analyzed the cellular and molecular mechanisms governing the adhesive and migratory behavior of enteric neural crest cells (ENCCs) during their collective migration within the developing mouse gut. We aimed to decipher the role of the complement anaphylatoxin C3a during this process, because this well-known immune system attractant has been implicated in cephalic NCC co-attraction, a process controlling directional migration. We used the conditional Ht-PA-cre transgenic mouse model allowing a specific ablation of the N-cadherin gene and the expression of a fluorescent reporter in migratory ENCCs without affecting the central nervous system. We performed time-lapse videomicroscopy of ENCCs from control and N-cadherin mutant gut explants cultured on fibronectin (FN) and micropatterned FN-stripes with C3a or C3aR antagonist, and studied cell migration behavior with the use of triangulation analysis to quantify cell dispersion. We performed ex vivo gut cultures with or without C3aR antagonist to determine the effect on ENCC behavior. Confocal microscopy was used to analyze the cell-matrix adhesion properties. We provide the first demonstration of the localization of the complement anaphylatoxin C3a and its receptor on ENCCs during their migration in the embryonic gut. C3aR receptor inhibition alters ENCC adhesion and migration, perturbing directionality and increasing cell dispersion both in vitro and ex vivo. N-cadherin-null ENCCs do not respond to C3a co-attraction. These findings indicate that C3a regulates cell migration in a N-cadherin-dependent process. Our results shed light on the role of C3a in regulating collective and directional cell migration, and in ganglia network organization during enteric nervous system ontogenesis. The detection of an immune system chemokine in ENCCs during ENS development may also shed light on new mechanisms for gastrointestinal disorders.
Keywords: Adhesion; Co-attraction; Complement anaphylatoxin C3a; Enteric nervous system; Migration; N-cadherin; Neural crest cells.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Abercrombie M, Heaysman JE. Observations on the social behaviour of cells in tissue culture. I. Speed of movement of chick heart fibroblasts in relation to their mutual contacts. Exp. Cell Res. 1953;5(1):111–131. - PubMed
-
- Abercrombie M, Heaysman JE. Observations on the social behaviour of cells in tissue culture. II. Monolayering of fibroblasts. Exp. Cell Res. 1954;6(2):293–306. - PubMed
-
- Ames RS, et al. Identification of a selective nonpeptide antagonist of the anaphylatoxin C3a receptor that demonstrates antiinflammatory activity in animal models. J. Immunol. 2001;166(10):6341–6348. - PubMed
-
- Anderson RB, et al. The cell adhesion molecule L1 Is required for chain migration of neural crest cells in the developing mouse gut. Gastroenterology. 2006;130(4):1221–1232. - PubMed
-
- Azioune A, et al. Simple and rapid process for single cell micro-patterning. Lab Chip. 2009;9(11):1640–1642. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

