Crystal structure of the human σ1 receptor
- PMID: 27042935
- PMCID: PMC5550834
- DOI: 10.1038/nature17391
Crystal structure of the human σ1 receptor
Abstract
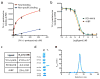
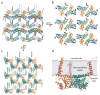
The human σ1 receptor is an enigmatic endoplasmic-reticulum-resident transmembrane protein implicated in a variety of disorders including depression, drug addiction, and neuropathic pain. Recently, an additional connection to amyotrophic lateral sclerosis has emerged from studies of human genetics and mouse models. Unlike many transmembrane receptors that belong to large, extensively studied families such as G-protein-coupled receptors or ligand-gated ion channels, the σ1 receptor is an evolutionary isolate with no discernible similarity to any other human protein. Despite its increasingly clear importance in human physiology and disease, the molecular architecture of the σ1 receptor and its regulation by drug-like compounds remain poorly defined. Here we report crystal structures of the human σ1 receptor in complex with two chemically divergent ligands, PD144418 and 4-IBP. The structures reveal a trimeric architecture with a single transmembrane domain in each protomer. The carboxy-terminal domain of the receptor shows an extensive flat, hydrophobic membrane-proximal surface, suggesting an intimate association with the cytosolic surface of the endoplasmic reticulum membrane in cells. This domain includes a cupin-like β-barrel with the ligand-binding site buried at its centre. This large, hydrophobic ligand-binding cavity shows remarkable plasticity in ligand recognition, binding the two ligands in similar positions despite dissimilar chemical structures. Taken together, these results reveal the overall architecture, oligomerization state, and molecular basis for ligand recognition by this important but poorly understood protein.
Conflict of interest statement
The authors declare no competing financial interests
Figures

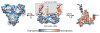







References
-
- Largent BL, Wikstrom H, Gundlach AL, Snyder SH. Structural determinants of sigma receptor affinity. Mol Pharmacol. 1987;32:772–784. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

