Decoding Polo-like kinase 1 signaling along the kinetochore-centromere axis
- PMID: 27043190
- PMCID: PMC4871769
- DOI: 10.1038/nchembio.2060
Decoding Polo-like kinase 1 signaling along the kinetochore-centromere axis
Abstract
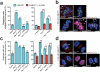
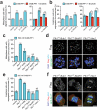
Protein kinase signaling along the kinetochore-centromere axis is crucial to assure mitotic fidelity, yet the details of its spatial coordination are obscure. Here, we examined how pools of human Polo-like kinase 1 (Plk1) within this axis control signaling events to elicit mitotic functions. To do this, we restricted active Plk1 to discrete subcompartments within the kinetochore-centromere axis using chemical genetics and decoded functional and phosphoproteomic signatures of each. We observe distinct phosphoproteomic and functional roles, suggesting that Plk1 exists and functions in discrete pools along this axis. Deep within the centromere, Plk1 operates to assure proper chromosome alignment and segregation. Thus, Plk1 at the kinetochore is a conglomerate of an observable bulk pool coupled with additional functional pools below the threshold of microscopic detection or resolution. Although complex, this multiplicity of locales provides an opportunity to decouple functional and phosphoproteomic signatures for a comprehensive understanding of Plk1's kinetochore functions.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

