Standardized benchmarking in the quest for orthologs
- PMID: 27043882
- PMCID: PMC4827703
- DOI: 10.1038/nmeth.3830
Standardized benchmarking in the quest for orthologs
Abstract
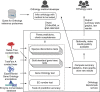
Achieving high accuracy in orthology inference is essential for many comparative, evolutionary and functional genomic analyses, yet the true evolutionary history of genes is generally unknown and orthologs are used for very different applications across phyla, requiring different precision-recall trade-offs. As a result, it is difficult to assess the performance of orthology inference methods. Here, we present a community effort to establish standards and an automated web-based service to facilitate orthology benchmarking. Using this service, we characterize 15 well-established inference methods and resources on a battery of 20 different benchmarks. Standardized benchmarking provides a way for users to identify the most effective methods for the problem at hand, sets a minimum requirement for new tools and resources, and guides the development of more accurate orthology inference methods.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Altenhoff, A.M. & Dessimoz, C. in Evolutionary Genomics (ed. Anisimova, M.) Ch. 9 (Humana Press, 2012).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

