DNA repair pathways underlie a common genetic mechanism modulating onset in polyglutamine diseases
- PMID: 27044000
- PMCID: PMC4914895
- DOI: 10.1002/ana.24656
DNA repair pathways underlie a common genetic mechanism modulating onset in polyglutamine diseases
Abstract
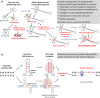
Objective: The polyglutamine diseases, including Huntington's disease (HD) and multiple spinocerebellar ataxias (SCAs), are among the commonest hereditary neurodegenerative diseases. They are caused by expanded CAG tracts, encoding glutamine, in different genes. Longer CAG repeat tracts are associated with earlier ages at onset, but this does not account for all of the difference, and the existence of additional genetic modifying factors has been suggested in these diseases. A recent genome-wide association study (GWAS) in HD found association between age at onset and genetic variants in DNA repair pathways, and we therefore tested whether the modifying effects of variants in DNA repair genes have wider effects in the polyglutamine diseases.
Methods: We assembled an independent cohort of 1,462 subjects with HD and polyglutamine SCAs, and genotyped single-nucleotide polymorphisms (SNPs) selected from the most significant hits in the HD study.
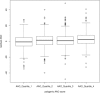
Results: In the analysis of DNA repair genes as a group, we found the most significant association with age at onset when grouping all polyglutamine diseases (HD+SCAs; p = 1.43 × 10(-5) ). In individual SNP analysis, we found significant associations for rs3512 in FAN1 with HD+SCAs (p = 1.52 × 10(-5) ) and all SCAs (p = 2.22 × 10(-4) ) and rs1805323 in PMS2 with HD+SCAs (p = 3.14 × 10(-5) ), all in the same direction as in the HD GWAS.
Interpretation: We show that DNA repair genes significantly modify age at onset in HD and SCAs, suggesting a common pathogenic mechanism, which could operate through the observed somatic expansion of repeats that can be modulated by genetic manipulation of DNA repair in disease models. This offers novel therapeutic opportunities in multiple diseases. Ann Neurol 2016;79:983-990.
© 2016 The Authors. Annals of Neurology published by Wiley Periodicals, Inc. on behalf of American Neurological Association.
Figures
References
-
- Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet 2005;6:743–755. - PubMed
-
- van de Warrenburg BP, Hendriks H, Durr A, et al. Age at onset variance analysis in spinocerebellar ataxias: a study in a Dutch‐French cohort. Ann Neurol 2005;57:505–512. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

