Functional network inference of the suprachiasmatic nucleus
- PMID: 27044085
- PMCID: PMC4843423
- DOI: 10.1073/pnas.1521178113
Functional network inference of the suprachiasmatic nucleus
Erratum in
-
Correction for Abel et al., Functional network inference of the suprachiasmatic nucleus.Proc Natl Acad Sci U S A. 2017 Oct 31;114(44):E9427. doi: 10.1073/pnas.1717182114. Epub 2017 Oct 23. Proc Natl Acad Sci U S A. 2017. PMID: 29078424 Free PMC article. No abstract available.
-
Correction to Supporting Information for Abel et al., Functional network inference of the suprachiasmatic nucleus.Proc Natl Acad Sci U S A. 2017 Oct 31;114(44):E9428-E9430. doi: 10.1073/pnas.1717183114. Epub 2017 Oct 23. Proc Natl Acad Sci U S A. 2017. PMID: 29078425 Free PMC article. No abstract available.
Abstract
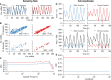
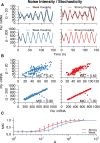
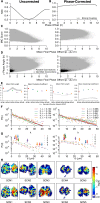
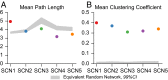
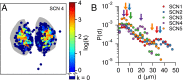
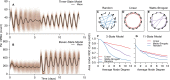
In the mammalian suprachiasmatic nucleus (SCN), noisy cellular oscillators communicate within a neuronal network to generate precise system-wide circadian rhythms. Although the intracellular genetic oscillator and intercellular biochemical coupling mechanisms have been examined previously, the network topology driving synchronization of the SCN has not been elucidated. This network has been particularly challenging to probe, due to its oscillatory components and slow coupling timescale. In this work, we investigated the SCN network at a single-cell resolution through a chemically induced desynchronization. We then inferred functional connections in the SCN by applying the maximal information coefficient statistic to bioluminescence reporter data from individual neurons while they resynchronized their circadian cycling. Our results demonstrate that the functional network of circadian cells associated with resynchronization has small-world characteristics, with a node degree distribution that is exponential. We show that hubs of this small-world network are preferentially located in the central SCN, with sparsely connected shells surrounding these cores. Finally, we used two computational models of circadian neurons to validate our predictions of network structure.
Keywords: biological clock; circadian oscillator; mathematical model; synchronization; systems biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Refinetti R, Menaker M. The circadian rhythm of body temperature. Physiol Behav. 1992;51(3):613–637. - PubMed
-
- Nagoshi E, et al. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119(5):693–705. - PubMed
-
- Sassone-Corsi P. Molecular clocks: Mastering time by gene regulation. Nature. 1998;392(6679):871–874. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

