Interallelic interaction and gene regulation in budding yeast
- PMID: 27044105
- PMCID: PMC4843442
- DOI: 10.1073/pnas.1601003113
Interallelic interaction and gene regulation in budding yeast
Abstract
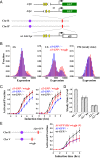
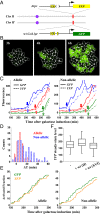
InDrosophila, homologous chromosome pairing leads to "transvection," in which the enhancer of a gene can regulate the allelic transcription intrans.Interallelic interactions were also observed in vegetative diploid budding yeast, but their functional significance is unknown. Here, we show that aGAL1reporter can interact with its homologous allele and affect its expression. By ectopically inserting two allelic reporters, one driven by wild-typeGAL1promoter (WTGAL1pr) and the other by a mutant promoter with delayed response to galactose induction, we found that the two reporters physically associate, and the WTGAL1prtriggers synchronized firing of the defective promoter and accelerates its activation without affecting its steady-state expression level. This interaction and the transregulatory effect disappear when the same reporters are located at nonallelic sites. We further demonstrated that the activator Gal4 is essential for the interallelic interaction, and the transregulation requires fully activated WTGAL1prtranscription. The mechanism of this phenomenon was further discussed. Taken together, our data revealed the existence of interallelic gene regulation in yeast, which serves as a starting point for understanding long-distance gene regulation in this genetically tractable system.
Keywords: homologous pairing; interallelic interaction; interallelic regulation; single-cell gene expression; transvection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
3D clustering of co-regulated genes and its effect on gene expression.Curr Genet. 2017 Dec;63(6):1017-1021. doi: 10.1007/s00294-017-0712-9. Epub 2017 May 27. Curr Genet. 2017. PMID: 28551816 Free PMC article. Review.
-
Bimodal expression of yeast GAL genes is controlled by a long non-coding RNA and a bifunctional galactokinase.Biochem Biophys Res Commun. 2017 Apr 22;486(1):63-69. doi: 10.1016/j.bbrc.2017.02.127. Epub 2017 Feb 27. Biochem Biophys Res Commun. 2017. PMID: 28254434
-
Galactose induction of the GAL1 gene requires conditional degradation of the Mig2 repressor.Biochem J. 2011 May 1;435(3):641-9. doi: 10.1042/BJ20102034. Biochem J. 2011. PMID: 21323640
-
A yeast catabolic enzyme controls transcriptional memory.Curr Biol. 2007 Dec 4;17(23):2041-6. doi: 10.1016/j.cub.2007.10.044. Epub 2007 Nov 8. Curr Biol. 2007. PMID: 17997309
-
Systems for applied gene control in Saccharomyces cerevisiae.Biotechnol Lett. 2008 Jun;30(6):979-87. doi: 10.1007/s10529-008-9647-z. Epub 2008 Feb 1. Biotechnol Lett. 2008. PMID: 18239858 Review.
Cited by
-
A combination of transcription factors mediates inducible interchromosomal contacts.Elife. 2019 May 13;8:e42499. doi: 10.7554/eLife.42499. Elife. 2019. PMID: 31081754 Free PMC article.
-
Interallelic Transcriptional Enhancement as an in Vivo Measure of Transvection in Drosophila melanogaster.G3 (Bethesda). 2016 Oct 13;6(10):3139-3148. doi: 10.1534/g3.116.032300. G3 (Bethesda). 2016. PMID: 27489208 Free PMC article.
-
The dynamic three-dimensional organization of the diploid yeast genome.Elife. 2017 May 24;6:e23623. doi: 10.7554/eLife.23623. Elife. 2017. PMID: 28537556 Free PMC article.
-
Subnuclear positioning and interchromosomal clustering of the GAL1-10 locus are controlled by separable, interdependent mechanisms.Mol Biol Cell. 2016 Oct 1;27(19):2980-93. doi: 10.1091/mbc.E16-03-0174. Epub 2016 Aug 3. Mol Biol Cell. 2016. PMID: 27489341 Free PMC article.
-
Three distinct mechanisms of long-distance modulation of gene expression in yeast.PLoS Genet. 2017 Apr 20;13(4):e1006736. doi: 10.1371/journal.pgen.1006736. eCollection 2017 Apr. PLoS Genet. 2017. PMID: 28426659 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

