Diversity of flux distribution in central carbon metabolism of S. cerevisiae strains from diverse environments
- PMID: 27044358
- PMCID: PMC4820951
- DOI: 10.1186/s12934-016-0456-0
Diversity of flux distribution in central carbon metabolism of S. cerevisiae strains from diverse environments
Abstract
Background: S. cerevisiae has attracted considerable interest in recent years as a model for ecology and evolutionary biology, revealing a substantial genetic and phenotypic diversity. However, there is a lack of knowledge on the diversity of metabolic networks within this species.
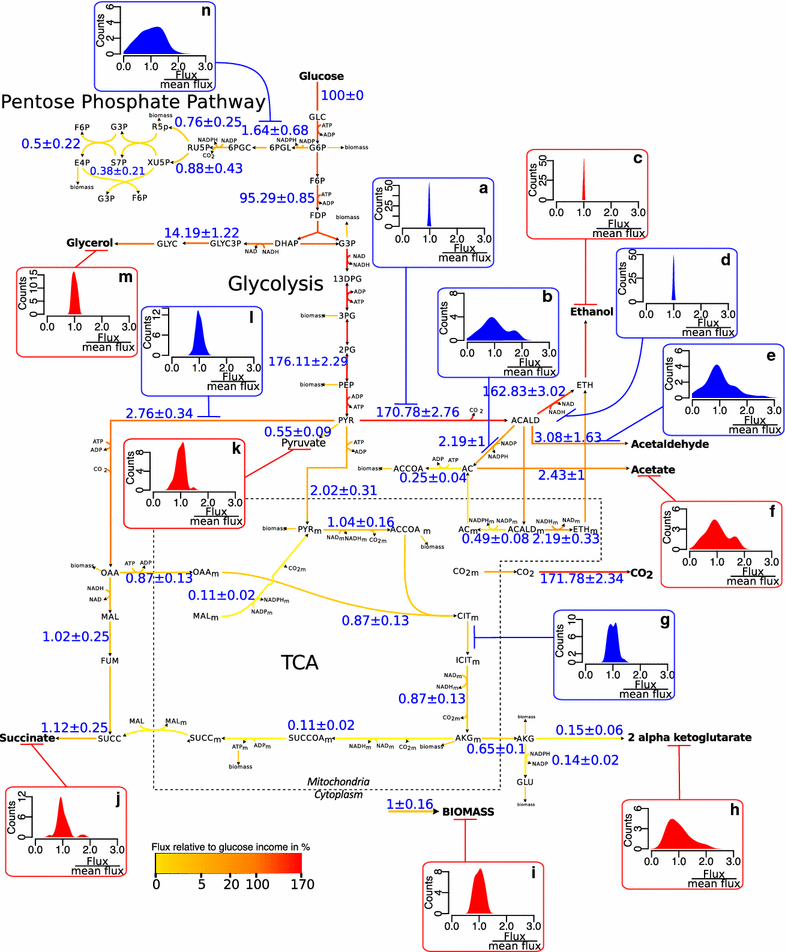
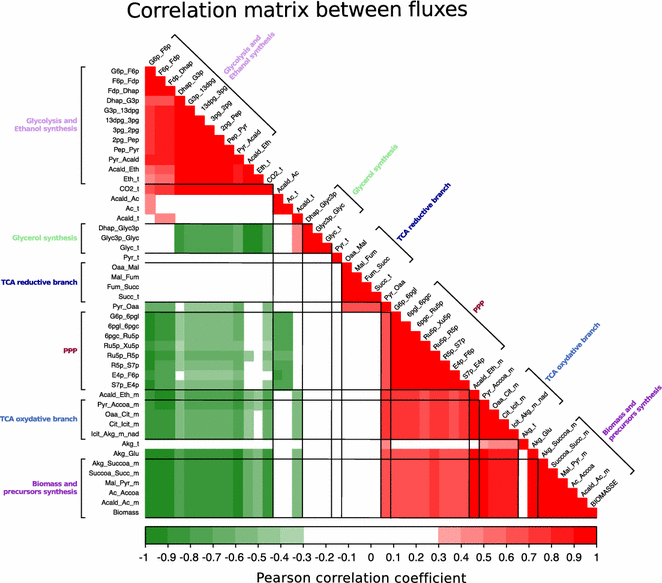
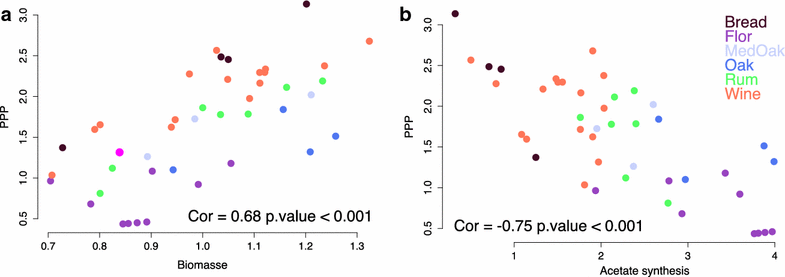
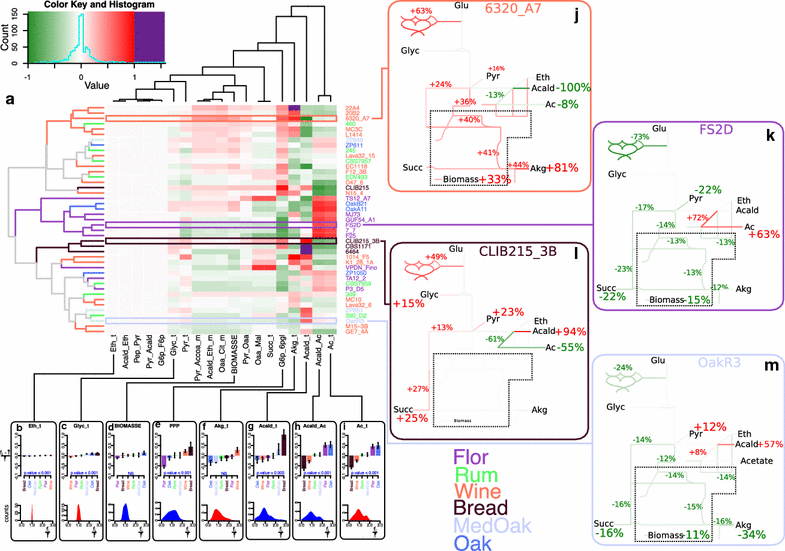
Results: To identify the metabolic and evolutionary constraints that shape metabolic fluxes in S. cerevisiae, we used a dedicated constraint-based model to predict the central carbon metabolism flux distribution of 43 strains from different ecological origins, grown in wine fermentation conditions. In analyzing these distributions, we observed a highly contrasted situation in flux variability, with quasi-constancy of the glycolysis and ethanol synthesis yield yet high flexibility of other fluxes, such as the pentose phosphate pathway and acetaldehyde production. Furthermore, these fluxes with large variability showed multimodal distributions that could be linked to strain origin, indicating a convergence between genetic origin and flux phenotype.
Conclusions: Flux variability is pathway-dependent and, for some flux, a strain origin effect can be found. These data highlight the constraints shaping the yeast operative central carbon network and provide clues for the design of strategies for strain improvement.
Keywords: Diversity; Flux balance analysis; Metabolic flux; Modeling; S. cerevisiae.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

