A general strategy for expanding polymerase function by droplet microfluidics
- PMID: 27044725
- PMCID: PMC4822039
- DOI: 10.1038/ncomms11235
A general strategy for expanding polymerase function by droplet microfluidics
Abstract
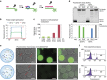
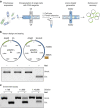
Polymerases that synthesize artificial genetic polymers hold great promise for advancing future applications in synthetic biology. However, engineering natural polymerases to replicate unnatural genetic polymers is a challenging problem. Here we present droplet-based optical polymerase sorting (DrOPS) as a general strategy for expanding polymerase function that employs an optical sensor to monitor polymerase activity inside the microenvironment of a uniform synthetic compartment generated by microfluidics. We validated this approach by performing a complete cycle of encapsulation, sorting and recovery on a doped library and observed an enrichment of ∼1,200-fold for a model engineered polymerase. We then applied our method to evolve a manganese-independent α-L-threofuranosyl nucleic acid (TNA) polymerase that functions with >99% template-copying fidelity. Based on our findings, we suggest that DrOPS is a versatile tool that could be used to evolve any polymerase function, where optical detection can be achieved by Watson-Crick base pairing.
Figures

References
-
- Yu H., Zhang S. & Chaput J. C. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat. Chem. 4, 183–187 (2012) . - PubMed
-
- Yu H., Zhang S., Dunn M. & Chaput J. C. An efficient and faithful in vitro replication system for threose nucleic acid. J. Am. Chem. Soc. 135, 3583–3591 (2013) . - PubMed
-
- Chaput J. C., Yu H. & Zhang S. The emerging world of synthetic genetics. Chem. Biol. 19, 1360–1371 (2012) . - PubMed
-
- Pinheiro V. B., Loakes D. & Holliger P. Synthetic polymers and their potential as genetic materials. Bioessays 35, 113–122 (2012) . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

