Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo
- PMID: 27044921
- PMCID: PMC5207318
- DOI: 10.15252/embj.201593360
Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo
Abstract
The molecular roles of many RNA-binding proteins in bacterial post-transcriptional gene regulation are not well understood. Approaches combining in vivo UV crosslinking with RNA deep sequencing (CLIP-seq) have begun to revolutionize the transcriptome-wide mapping of eukaryotic RNA-binding protein target sites. We have applied CLIP-seq to chart the target landscape of two major bacterial post-transcriptional regulators, Hfq and CsrA, in the model pathogen Salmonella Typhimurium. By detecting binding sites at single-nucleotide resolution, we identify RNA preferences and structural constraints of Hfq and CsrA during their interactions with hundreds of cellular transcripts. This reveals 3'-located Rho-independent terminators as a universal motif involved in Hfq-RNA interactions. Additionally, Hfq preferentially binds 5' to sRNA-target sites in mRNAs, and 3' to seed sequences in sRNAs, reflecting a simple logic in how Hfq facilitates sRNA-mRNA interactions. Importantly, global knowledge of Hfq sites significantly improves sRNA-target predictions. CsrA binds AUGGA sequences in apical loops and targets many Salmonella virulence mRNAs. Overall, our generic CLIP-seq approach will bring new insights into post-transcriptional gene regulation by RNA-binding proteins in diverse bacterial species.
Keywords: CLIP; CsrA; Hfq; non‐coding RNA; peak calling; post‐transcriptional control; small RNA; terminator; translation.
© 2016 The Authors. Published under the terms of the CC BY NC ND 4.0 license.
Figures
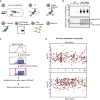
Schematic representation of the CLIP‐seq protocol for bacterial RBPs that was established and used in this study. UV: ultraviolet.
Detection of crosslinked, immunoprecipitated, and radioactively labeled RNA–protein complexes after separation on denaturing SDS–polyacrylamide gels and transfer to nitrocellulose membranes. Radioactive signals were detected by phosphorimaging (top). Detection of Hfq‐3xFLAG proteins by Western blot using an anti‐FLAG antibody served as a control for successful immunoprecipitation (bottom). CL: crosslinking.
Schematic representation of binding site determination (peak calling).
Fold change (y‐axis) and genomic position (x‐axis) of Hfq peaks. Mbp: mega basepair.
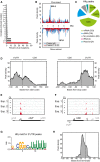
- A
Percentage of the occurrence of the indicated mutations among all crosslink‐specific mutations found within Hfq peaks.
- B
Hfq peak distribution along the Salmonella chromosome divided in bins of 2 × 104 basepairs each. The genomic positions of the pathogenicity islands SPI‐1 and SPI‐2 are indicated. Mbp: mega basepair.
- C
Distribution of Hfq peaks among the indicated RNA classes. Numbers in parentheses give the number of called peaks that overlapped with annotations belonging to the respective RNA class.
- D
Global peak density distribution (meta‐gene analysis) around start and stop codons. For this analysis, only those start and stop codons were used that are flanked by a 5′UTR or 3′UTR, respectively. Vertical dashed lines indicate the position of start and stop codons, respectively.
- E, F
Read coverage at the chiP (E) and hilD (F) loci in libraries from crosslinked and non‐crosslinked samples. Exp: experiment, CL: crosslinking
- G
Consensus motif generated by MEME using sequences of Hfq peaks mapping to mRNA 3′UTRs.
- H
Meta‐gene analysis of peak distribution around genomic positions of predicted Rho‐independent terminators.


Read coverage in libraries from crosslinked and non‐crosslinked samples at the sgrS locus. CL: crosslinking
Predicted secondary structure of the sRNA SgrS. Nucleotides corresponding to a Hfq peak and positions of crosslink‐induced mutations are color coded as highlighted in the legend.
Read coverage in libraries from crosslinked and non‐crosslinked samples at the rydC locus. CL: crosslinking.
Predicted pseudoknot structure of the sRNA RydC. Nucleotides corresponding to an Hfq peak and positions of crosslink‐induced mutations are color coded as highlighted in (B).
Meta‐gene analysis of the peak distribution along Salmonella sRNAs. Length normalization was achieved through proportional binning according to the different lengths of the sRNA sequences.
Consensus motif generated by MEME using sequences of peaks mapping to sRNAs as input.
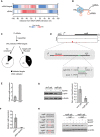
Distribution of Hfq peaks with respect to sRNA interaction sites in mRNA targets and seed sequences in sRNAs, respectively.
Putative model of Hfq interaction with cognate sRNA–mRNA pairs.
Workflow for the integration of Hfq peak information during sRNA‐target prediction using CopraRNA. The pie charts show the number of previously validated targets among all predictions, or among predicted targets with Hfq peaks, respectively.
Read coverage from Hfq CLIP‐seq at the mglB locus (top), location of the detected Hfq peak (red) and the predicted Spot42 interaction site (green) in the mglB 5′UTR (middle), and the predicted basepair interaction between Spot42 and mglB (bottom). The Spot42 interaction site in mglB is highlighted in green.
qRT–PCR analysis of mglB mRNA expression in wt Salmonella or in an isogenic Δspf strain. Samples were collected from cells grown in LB medium to an optical density of 0.3 (OD600). Means and error bars representing standard deviations are based on two biological replicates.
qRT–PCR analysis of mglB mRNA expression in Salmonella Δspf 10 min after induction of Spot42 overexpression from plasmid pBAD–Spot42. Plasmid pBAD was used as a control. Means and error bars representing standard deviations are based on two biological replicates.
Western blot analysis of GFP expression from plasmid‐expressed translational lacZ‐gfp and mglB‐gfp fusions in the presence or absence of Spot42 overexpression. Quantification of Western blot signals is shown on the right. Means and error bars representing standard deviations are based on three biological replicates. GFP fusion proteins were detected with an anti‐GFP antibody, while an anti‐GroEL antibody was used to determine the amount of protein loaded on the gel.
Western blot analysis of GFP expression from the wild‐type mglB‐gfp or mutant mglB*‐gfp fusions upon deletion and overexpression of wild‐type Spot42 or the Spot42* mutant. The predicted interactions between Spot42 and mglB, as well as the introduced mutations, are shown.

Putative feed‐forward loop between CRP‐cAMP, Spot42, and mglB.
Conservation of the predicted Spot42‐binding site in mglB mRNA. Sequence alignment of RNA sequences upstream of the mglB start codon. Gray shading highlights the predicted Spot42‐binding site. The alignment was made using MAFFT (Katoh et al, 2002). An asterisk indicates nucleotides that are identical in all sequences. ECO: Escherichia coli MG1655, CKO: Citrobacter koseri, CRO: Citrobacter rodentium, STM: Salmonella Typhimurium LT2, ENT: Enterobacter sp. 638, SPR: Serratia proteamaculans, YEN: Yersinia enterocolitica 8081.

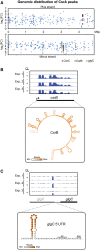
Fold change (y‐axis) and genomic position (x‐axis) of CsrA peaks. Peaks mapping to the known CsrA ligands CsrB, CsrC, and glgC are indicated.
Read coverage from CsrA CLIP‐seq at the csrB locus (top). A heat map of the average read coverage at the csrB locus superimposed on the predicted secondary structure of Salmonella CsrB (bottom). The CsrB structure was predicted by MFOLD (Zuker, 2003).
Read coverage from CsrA CLIP‐seq at the glgC locus (top). A heat map of the average read coverage at the glgC locus superimposed on the predicted secondary structure of the 5′UTR of the Salmonella glgC mRNA (bottom).

Distribution of CsrA peaks among the indicated RNA classes. Numbers in parenthesis represent the number of called peaks that were mapped within annotations belonging to the respective RNA class.
Meta‐gene analysis of CsrA peaks around start and stop codons. For this analysis, only those start and stop codons were used that are flanked by a 5′UTR or 3′UTR, respectively.
Percentage of peaks that contain the indicated sequences.
Consensus motif generated by MEME based on all CsrA peak sequences.
Percentage of the occurrence of the indicated mutations among all crosslink‐specific mutations found within CsrA peaks. The inset shows the consensus motif generated with MEME using sequences flanking a crosslink‐specific T to C mutation as input.
Consensus motifs generated by CMfinder based on all CsrA peaks.

CsrA peak density distribution along the Salmonella chromosome in bins of 2 × 104 basepairs. The genomic positions of Salmonella pathogenicity islands SPI‐1 and SPI‐2 are indicated.
KEGG pathways that were found significantly enriched among gene annotations to which CsrA peaks were mapped. Pathways that are related to Salmonella pathogenicity are highlighted in red.
Read coverage from CsrA CLIP‐seq at the sopD2 locus. Light blue bars represent called peaks.
Western blot analysis of SopD2‐GFP expression from a translational sopD2‐gfp fusion on a plasmid in the indicated strain backgrounds. Plus sign indicates the presence of plasmid pCsrB. Minus sign indicates the presence of the control vector pJV300. SopD2‐GFP signals were detected with an anti‐GFP antibody. Expression of GroEL served as a loading control and was detected with an anti‐GroEL antibody.
Predicted secondary structure of the sopD2 5′UTR. Peak position, GGA motifs, and introduced mutations are indicated. GFP fluorescence measurements from the wild‐type sopD2‐gfp fusion or a 2xCCU mutant upon csrBcsrC deletion and CsrB complementation. Means and error bars representing standard deviations are based on three independent experiments.
Read coverage at the prgHIJK‐orgAB locus from a CsrA CLIP‐seq experiment.
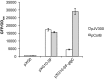
Western blot analysis of the expression from the indicated plasmid‐borne translational GFP fusions in the presence of plasmids pCsrB (plus signs) or pJV300 (minus signs).
Read coverage at the sicA‐sipBCDA‐iacP locus from a CsrA CLIP‐seq experiment.
Western blot analysis of the expression from the indicated plasmid‐borne translational GFP fusions in the presence of plasmids pCsrB (plus signs) or pJV300 (minus signs).

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

