Protruding Features of Viral Capsids Are Clustered on Icosahedral Great Circles
- PMID: 27045511
- PMCID: PMC4821576
- DOI: 10.1371/journal.pone.0152319
Protruding Features of Viral Capsids Are Clustered on Icosahedral Great Circles
Abstract
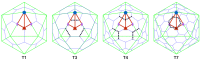
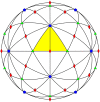
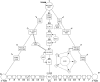
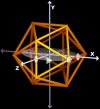
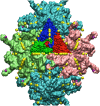
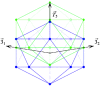
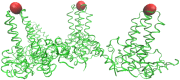
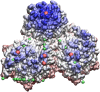
Spherical viruses are remarkably well characterized by the Triangulation (T) number developed by Casper and Klug. The T-number specifies how many viral capsid proteins are required to cover the virus, as well as how they are further subdivided into pentamer and hexamer subunits. The T-number however does not constrain the orientations of these proteins within the subunits or dictate where the proteins should place their protruding features. These protrusions often take the form of loops, spires and helices, and are significant because they aid in stability of the capsid as well as recognition by the host organism. Until now there has be no overall understanding of the placement of protrusions for spherical viruses, other than they have icosahedral symmetry. We constructed a set of gauge points based upon the work affine extensions of Keef and Twarock, which have fixed relative angular locations with which to measure the locations of these features. This work adds a new element to our understanding of the geometric arrangement of spherical viral capsid proteins; chiefly that the locations of protruding features are not found stochastically distributed in an icosahedral manner across the viral surface, but instead these features are found only in specific locations along the 15 icosahedral great circles. We have found that this result holds true as the T number and viral capsids size increases, suggesting an underlying geometric constraint on their locations. This is in spite of the fact that the constraints on the pentamers and hexamer orientations change as a function of T-number, as you need to accommodate more hexamers in the same solid angle between pentamers. The existence of this angular constraint of viral capsids suggests that there is a fitness or energetic benefit to the virus placing its protrusions in this manner. This discovery may have profound impacts on identifying and eliminating viral pathogens, understanding evolutionary constraints as well as bioengineering for capsid drug delivery systems. This result also suggests that in addition to biochemical attachment restrictions, there are additional geometric constraints that should be adhered to when modifying protein capsids.
Conflict of interest statement
Figures





Similar articles
-
Unveiling the Hidden Rules of Spherical Viruses Using Point Arrays.Viruses. 2020 Apr 20;12(4):467. doi: 10.3390/v12040467. Viruses. 2020. PMID: 32326043 Free PMC article.
-
Viral Phrenology.Viruses. 2021 Oct 30;13(11):2191. doi: 10.3390/v13112191. Viruses. 2021. PMID: 34834999 Free PMC article.
-
Affine extensions of the icosahedral group with applications to the three-dimensional organisation of simple viruses.J Math Biol. 2009 Sep;59(3):287-313. doi: 10.1007/s00285-008-0228-5. Epub 2008 Nov 1. J Math Biol. 2009. PMID: 18979101
-
Breaking Symmetry in Viral Icosahedral Capsids as Seen through the Lenses of X-ray Crystallography and Cryo-Electron Microscopy.Viruses. 2018 Feb 7;10(2):67. doi: 10.3390/v10020067. Viruses. 2018. PMID: 29414851 Free PMC article. Review.
-
Geometric architecture of viruses.World J Virol. 2020 Aug 25;9(2):5-18. doi: 10.5501/wjv.v9.i2.5. World J Virol. 2020. PMID: 32923381 Free PMC article. Review.
Cited by
-
A Capsid Structure of Ralstonia solanacearum podoviridae GP4 with a Triangulation Number T = 9.Viruses. 2022 Nov 1;14(11):2431. doi: 10.3390/v14112431. Viruses. 2022. PMID: 36366529 Free PMC article.
-
Keeping It Together: Structures, Functions, and Applications of Viral Decoration Proteins.Viruses. 2020 Oct 14;12(10):1163. doi: 10.3390/v12101163. Viruses. 2020. PMID: 33066635 Free PMC article. Review.
-
Unveiling the Hidden Rules of Spherical Viruses Using Point Arrays.Viruses. 2020 Apr 20;12(4):467. doi: 10.3390/v12040467. Viruses. 2020. PMID: 32326043 Free PMC article.
-
Plant-Derived Epi-Nutraceuticals as Potential Broad-Spectrum Anti-Viral Agents.Nutrients. 2023 Nov 8;15(22):4719. doi: 10.3390/nu15224719. Nutrients. 2023. PMID: 38004113 Free PMC article. Review.
-
Viral Phrenology.Viruses. 2021 Oct 30;13(11):2191. doi: 10.3390/v13112191. Viruses. 2021. PMID: 34834999 Free PMC article.
References
-
- Hubert H, Devouard B, Garvie L. Icosahedral packing of B12 icosahedra in boron suboxide (B6O). Nature. 1998;391(January):376–378. 10.1038/34885 - DOI
-
- Carrillo-Tripp M, Shepherd CM, Borelli Ia, Venkataraman S, Lander G, Natarajan P, et al. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic acids research. 2009. January;37(Database issue):D436–42. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2686430&tool=p... 10.1093/nar/gkn840 - DOI - PMC - PubMed
-
- Sällberg M, Rudén U, Magnius LO, Harthus HP, Noah M, Wahren B. Characterisation of a linear binding site for a monoclonal antibody to hepatitis B core antigen. Journal of medical virology. 1991. April;33(4):248–52. Available from: http://www.ncbi.nlm.nih.gov/pubmed/1713264 10.1002/jmv.1890330407 - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

