Lysosomal putative RNA transporter SIDT2 mediates direct uptake of RNA by lysosomes
- PMID: 27046251
- PMCID: PMC4836006
- DOI: 10.1080/15548627.2016.1145325
Lysosomal putative RNA transporter SIDT2 mediates direct uptake of RNA by lysosomes
Abstract
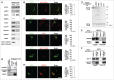
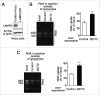
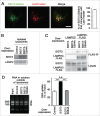
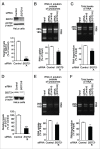
Lysosomes are thought to be the major intracellular compartment for the degradation of macromolecules. We recently identified a novel type of autophagy, RNautophagy, where RNA is directly taken up by lysosomes in an ATP-dependent manner and degraded. However, the mechanism of RNA translocation across the lysosomal membrane and the physiological role of RNautophagy remain unclear. In the present study, we performed gain- and loss-of-function studies with isolated lysosomes, and found that SIDT2 (SID1 transmembrane family, member 2), an ortholog of the Caenorhabditis elegans putative RNA transporter SID-1 (systemic RNA interference deficient-1), mediates RNA translocation during RNautophagy. We also observed that SIDT2 is a transmembrane protein, which predominantly localizes to lysosomes. Strikingly, knockdown of Sidt2 inhibited up to ˜50% of total RNA degradation at the cellular level, independently of macroautophagy. Moreover, we showed that this impairment is mainly due to inhibition of lysosomal RNA degradation, strongly suggesting that RNautophagy plays a significant role in constitutive cellular RNA degradation. Our results provide a novel insight into the mechanisms of RNA metabolism, intracellular RNA transport, and atypical types of autophagy.
Keywords: RNA; RNautophagy; autophagy; lysosome; membrane protein.
Figures


References
-
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147:728-41; PMID:22078875; http://dx.doi.org/ 10.1016/j.cell.2011.10.026 - DOI - PubMed
-
- Kaushik S, Cuervo AM. Methods to monitor chaperone-mediated autophagy. Methods Enzymol 2009; 452:297-324; PMID:19200890; http://dx.doi.org/ 10.1016/S0076-6879(08)03619-7 - DOI - PMC - PubMed
-
- Fujiwara Y, Furuta A, Kikuchi H, Aizawa S, Hatanaka Y, Konya C, Uchida K, Yoshimura A, Tamai Y, Wada K, et al.. Discovery of a novel type of autophagy targeting RNA. Autophagy 2013; 9:403-9; PMID:23291500; http://dx.doi.org/ 10.4161/auto.23002 - DOI - PMC - PubMed
-
- Fujiwara Y, Kikuchi H, Aizawa S, Furuta A, Hatanaka Y, Konya C, Uchida K, Wada K, Kabuta T. Direct uptake and degradation of DNA by lysosomes. Autophagy 2013; 9:1167-71; http://dx.doi.org/ 10.4161/auto.24880 - DOI - PMC - PubMed
-
- Fujiwara Y, Hase K, Wada K, Kabuta T. An RNautophagy/DNautophagy receptor, LAMP2C, possesses an arginine-rich motif that mediates RNA/DNA-binding. Biochem Biophys Res Commun 2015; 460:281-6; PMID:25772617; http://dx.doi.org/ 10.1016/j.bbrc.2015.03.025 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
