Data- and knowledge-based modeling of gene regulatory networks: an update
- PMID: 27047314
- PMCID: PMC4817425
- DOI: 10.17179/excli2015-168
Data- and knowledge-based modeling of gene regulatory networks: an update
Abstract
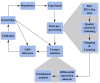
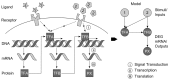
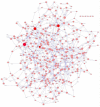
Gene regulatory network inference is a systems biology approach which predicts interactions between genes with the help of high-throughput data. In this review, we present current and updated network inference methods focusing on novel techniques for data acquisition, network inference assessment, network inference for interacting species and the integration of prior knowledge. After the advance of Next-Generation-Sequencing of cDNAs derived from RNA samples (RNA-Seq) we discuss in detail its application to network inference. Furthermore, we present progress for large-scale or even full-genomic network inference as well as for small-scale condensed network inference and review advances in the evaluation of network inference methods by crowdsourcing. Finally, we reflect the current availability of data and prior knowledge sources and give an outlook for the inference of gene regulatory networks that reflect interacting species, in particular pathogen-host interactions.
Keywords: RNA-Seq; gene regulatory networks; modeling; network inference; prior knowledge; reverse engineering.
Figures

Similar articles
-
How to Predict Molecular Interactions between Species?Front Microbiol. 2016 Mar 31;7:442. doi: 10.3389/fmicb.2016.00442. eCollection 2016. Front Microbiol. 2016. PMID: 27065992 Free PMC article. Review.
-
An algebra-based method for inferring gene regulatory networks.BMC Syst Biol. 2014 Mar 26;8:37. doi: 10.1186/1752-0509-8-37. BMC Syst Biol. 2014. PMID: 24669835 Free PMC article.
-
Gene expression complex networks: synthesis, identification, and analysis.J Comput Biol. 2011 Oct;18(10):1353-67. doi: 10.1089/cmb.2010.0118. Epub 2011 May 6. J Comput Biol. 2011. PMID: 21548810
-
Gene regulatory network inference: evaluation and application to ovarian cancer allows the prioritization of drug targets.Genome Med. 2012 May 1;4(5):41. doi: 10.1186/gm340. Genome Med. 2012. PMID: 22548828 Free PMC article.
-
Computational inference of gene regulatory networks: Approaches, limitations and opportunities.Biochim Biophys Acta Gene Regul Mech. 2017 Jan;1860(1):41-52. doi: 10.1016/j.bbagrm.2016.09.003. Epub 2016 Sep 16. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 27641093 Review.
Cited by
-
A review on computational systems biology of pathogen-host interactions.Front Microbiol. 2015 Apr 9;6:235. doi: 10.3389/fmicb.2015.00235. eCollection 2015. Front Microbiol. 2015. PMID: 25914674 Free PMC article. Review.
-
A strategy to incorporate prior knowledge into correlation network cutoff selection.Nat Commun. 2020 Oct 14;11(1):5153. doi: 10.1038/s41467-020-18675-3. Nat Commun. 2020. PMID: 33056991 Free PMC article.
-
How to Predict Molecular Interactions between Species?Front Microbiol. 2016 Mar 31;7:442. doi: 10.3389/fmicb.2016.00442. eCollection 2016. Front Microbiol. 2016. PMID: 27065992 Free PMC article. Review.
-
Reconstruction of the temporal signaling network in Salmonella-infected human cells.Front Microbiol. 2015 Jul 20;6:730. doi: 10.3389/fmicb.2015.00730. eCollection 2015. Front Microbiol. 2015. PMID: 26257716 Free PMC article.
-
JacLy: a Jacobian-based method for the inference of metabolic interactions from the covariance of steady-state metabolome data.PeerJ. 2018 Dec 6;6:e6034. doi: 10.7717/peerj.6034. eCollection 2018. PeerJ. 2018. PMID: 30564518 Free PMC article.
References
-
- Äijö T, Lähdesmäki H. Learning gene regulatory networks from gene expression measurements using non-parametric molecular kinetics. Bioinformatics. 2009;25:2937–2944. - PubMed
-
- Akutsu T, Miyano S, Kuhara S. Identification of genetic networks from a small number of gene expression patterns under the Boolean network model. Pac Symp Biocomput. 1999:17–28. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
