The potential of single-cell profiling in plants
- PMID: 27048384
- PMCID: PMC4820866
- DOI: 10.1186/s13059-016-0931-2
The potential of single-cell profiling in plants
Abstract
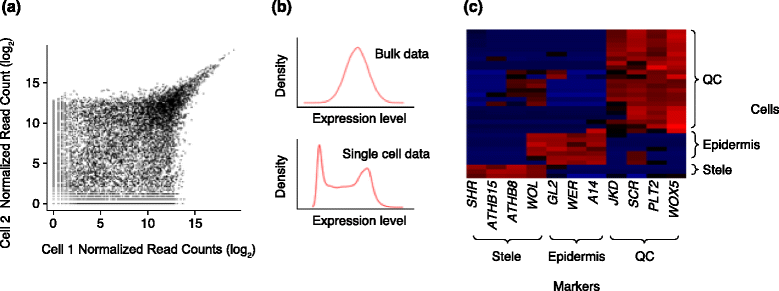
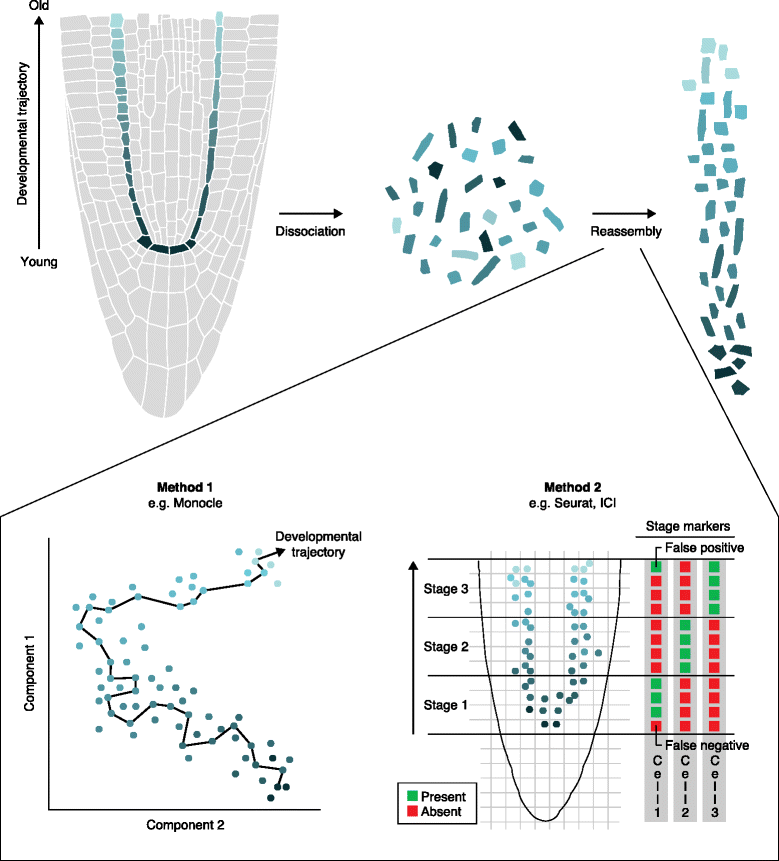
Single-cell transcriptomics has been employed in a growing number of animal studies, but the technique has yet to be widely used in plants. Nonetheless, early studies indicate that single-cell RNA-seq protocols developed for animal cells produce informative datasets in plants. We argue that single-cell transcriptomics has the potential to provide a new perspective on plant problems, such as the nature of the stem cells or initials, the plasticity of plant cells, and the extent of localized cellular responses to environmental inputs. Single-cell experimental outputs require different analytical approaches compared with pooled cell profiles and new tools tailored to single-cell assays are being developed. Here, we highlight promising new single-cell profiling approaches, their limitations as applied to plants, and their potential to address fundamental questions in plant biology.
Figures
Similar articles
-
Single-Cell Transcriptomics Applied in Plants.Cells. 2024 Sep 17;13(18):1561. doi: 10.3390/cells13181561. Cells. 2024. PMID: 39329745 Free PMC article. Review.
-
Designing a transcriptome next-generation sequencing project for a nonmodel plant species.Am J Bot. 2012 Feb;99(2):257-66. doi: 10.3732/ajb.1100292. Epub 2012 Jan 19. Am J Bot. 2012. PMID: 22268224 Review.
-
Scalable microfluidics for single-cell RNA printing and sequencing.Genome Biol. 2015 Jun 6;16(1):120. doi: 10.1186/s13059-015-0684-3. Genome Biol. 2015. PMID: 26047807 Free PMC article.
-
Single-cell transcriptomics: a new frontier in plant biotechnology research.Plant Cell Rep. 2024 Nov 25;43(12):294. doi: 10.1007/s00299-024-03383-9. Plant Cell Rep. 2024. PMID: 39585480 Review.
-
Quantitative single-cell transcriptomics.Brief Funct Genomics. 2018 Jul 1;17(4):220-232. doi: 10.1093/bfgp/ely009. Brief Funct Genomics. 2018. PMID: 29579145 Free PMC article. Review.
Cited by
-
Single-cell analysis of cell identity in the Arabidopsis root apical meristem: insights and opportunities.J Exp Bot. 2021 Oct 13;72(19):6679-6686. doi: 10.1093/jxb/erab228. J Exp Bot. 2021. PMID: 34018001 Free PMC article. Review.
-
Whole-genomic DNA amplifications from individually isolated sweet sorghum microspores.Appl Plant Sci. 2022 Nov 22;10(6):e11501. doi: 10.1002/aps3.11501. eCollection 2022 Nov-Dec. Appl Plant Sci. 2022. PMID: 36518943 Free PMC article.
-
Defining Cell Identity with Single-Cell Omics.Proteomics. 2018 Sep;18(18):e1700312. doi: 10.1002/pmic.201700312. Epub 2018 May 28. Proteomics. 2018. PMID: 29644800 Free PMC article. Review.
-
Laser Microdissection of Specific Stem-Base Tissue Types from Olive Microcuttings for Isolation of High-Quality RNA.Biology (Basel). 2021 Mar 10;10(3):209. doi: 10.3390/biology10030209. Biology (Basel). 2021. PMID: 33801829 Free PMC article.
-
Statistical and Machine Learning Approaches to Predict Gene Regulatory Networks From Transcriptome Datasets.Front Plant Sci. 2018 Nov 29;9:1770. doi: 10.3389/fpls.2018.01770. eCollection 2018. Front Plant Sci. 2018. PMID: 30555503 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

