Conservation and divergence of the histone code in nucleomorphs
- PMID: 27048461
- PMCID: PMC4822330
- DOI: 10.1186/s13062-016-0119-4
Conservation and divergence of the histone code in nucleomorphs
Abstract
Background: Nucleomorphs, the remnant nuclei of photosynthetic algae that have become endosymbionts to other eukaryotes, represent a unique example of convergent reductive genome evolution in eukaryotes, having evolved independently on two separate occasions in chlorarachniophytes and cryptophytes. The nucleomorphs of the two groups have evolved in a remarkably convergent manner, with numerous very similar features. Chief among them is the extreme reduction and compaction of nucleomorph genomes, with very small chromosomes and extremely short or even completely absent intergenic spaces. These characteristics pose a number of intriguing questions regarding the mechanisms of transcription and gene regulation in such a crowded genomic context, in particular in terms of the functioning of the histone code, which is common to almost all eukaryotes and plays a central role in chromatin biology.
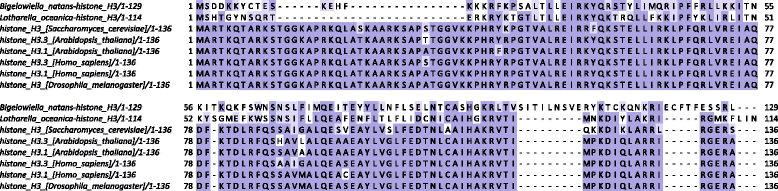
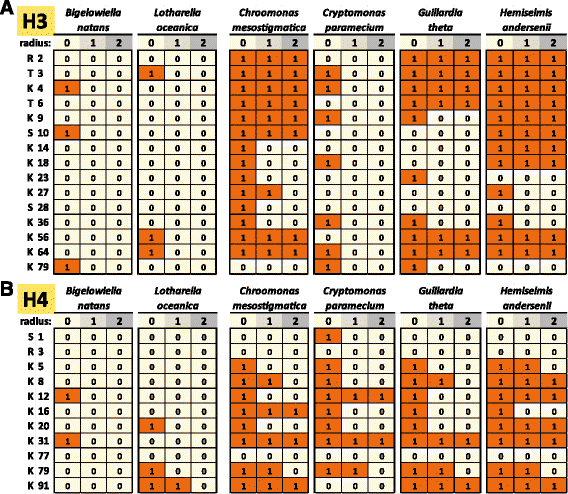
Results: This study examines the sequences of nucleomorph histone proteins in order to address these issues. Remarkably, all classical transcription- and repression-related components of the histone code seem to be missing from chlorarachniophyte nucleomorphs. Cryptophyte nucleomorph histones are generally more similar to the conventional eukaryotic state; however, they also display significant deviations from the typical histone code. Based on the analysis of specific components of the code, we discuss the state of chromatin and the transcriptional machinery in these nuclei.
Conclusions: The results presented here shed new light on the mechanisms of nucleomorph transcription and gene regulation and provide a foundation for future studies of nucleomorph chromatin and transcriptional biology.
Keywords: Chromatin; Histone code; Histones; Nucleomorphs; Transcription.
Figures



Similar articles
-
The chromatin organization of a chlorarachniophyte nucleomorph genome.Genome Biol. 2022 Mar 1;23(1):65. doi: 10.1186/s13059-022-02639-5. Genome Biol. 2022. PMID: 35232465 Free PMC article.
-
Nucleomorph and plastid genome sequences of the chlorarachniophyte Lotharella oceanica: convergent reductive evolution and frequent recombination in nucleomorph-bearing algae.BMC Genomics. 2014 May 15;15(1):374. doi: 10.1186/1471-2164-15-374. BMC Genomics. 2014. PMID: 24885563 Free PMC article.
-
Going, going, not quite gone: nucleomorphs as a case study in nuclear genome reduction.J Hered. 2009 Sep-Oct;100(5):582-90. doi: 10.1093/jhered/esp055. Epub 2009 Jul 17. J Hered. 2009. PMID: 19617523 Review.
-
Nucleomorph genomes.Annu Rev Genet. 2009;43:251-64. doi: 10.1146/annurev-genet-102108-134809. Annu Rev Genet. 2009. PMID: 19686079 Review.
-
Nucleomorph genome of Hemiselmis andersenii reveals complete intron loss and compaction as a driver of protein structure and function.Proc Natl Acad Sci U S A. 2007 Dec 11;104(50):19908-13. doi: 10.1073/pnas.0707419104. Epub 2007 Dec 6. Proc Natl Acad Sci U S A. 2007. PMID: 18077423 Free PMC article.
Cited by
-
Peculiarities of Plasmodium falciparum Gene Regulation and Chromatin Structure.Int J Mol Sci. 2021 May 13;22(10):5168. doi: 10.3390/ijms22105168. Int J Mol Sci. 2021. PMID: 34068393 Free PMC article. Review.
-
The chromatin landscape of the euryarchaeon Haloferax volcanii.Genome Biol. 2023 Nov 6;24(1):253. doi: 10.1186/s13059-023-03095-5. Genome Biol. 2023. PMID: 37932847 Free PMC article.
-
Relative Mutation Rates in Nucleomorph-Bearing Algae.Genome Biol Evol. 2019 Apr 1;11(4):1045-1053. doi: 10.1093/gbe/evz056. Genome Biol Evol. 2019. PMID: 30859201 Free PMC article.
-
The chromatin organization of a chlorarachniophyte nucleomorph genome.Genome Biol. 2022 Mar 1;23(1):65. doi: 10.1186/s13059-022-02639-5. Genome Biol. 2022. PMID: 35232465 Free PMC article.
-
The iTRAQ-based chloroplast proteomic analysis of Triticum aestivum L. leaves subjected to drought stress and 5-aminolevulinic acid alleviation reveals several proteins involved in the protection of photosynthesis.BMC Plant Biol. 2020 Mar 4;20(1):96. doi: 10.1186/s12870-020-2297-6. BMC Plant Biol. 2020. PMID: 32131734 Free PMC article.
References
-
- Greenwood AD. The Cryptophyta in relation to phylogeny and photosynthesis. In: Sanders JV, Goodchild DJ, editors. Electron microscopy 1974. Canberra: Australian Academy of Sciences; 1974.
-
- Greenwood AD, Griffiths HB, Santore UJ. Chloroplasts and cell compartments in Cryptophyceae. Brit Phycol J. 1977;12:119.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

