Computational analysis of EBNA1 "druggability" suggests novel insights for Epstein-Barr virus inhibitor design
- PMID: 27048620
- PMCID: PMC5180362
- DOI: 10.1007/s10822-016-9899-y
Computational analysis of EBNA1 "druggability" suggests novel insights for Epstein-Barr virus inhibitor design
Abstract
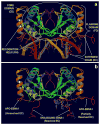
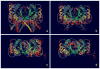
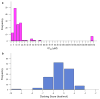
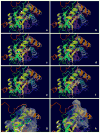
The Epstein-Barr Nuclear Antigen 1 (EBNA1) is a critical protein encoded by the Epstein-Barr Virus (EBV). During latent infection, EBNA1 is essential for DNA replication and transcription initiation of viral and cellular genes and is necessary to immortalize primary B-lymphocytes. Nonetheless, the concept of EBNA1 as drug target is novel. Two EBNA1 crystal structures are publicly available and the first small-molecule EBNA1 inhibitors were recently discovered. However, no systematic studies have been reported on the structural details of EBNA1 "druggable" binding sites. We conducted computational identification and structural characterization of EBNA1 binding pockets, likely to accommodate ligand molecules (i.e. "druggable" binding sites). Then, we validated our predictions by docking against a set of compounds previously tested in vitro for EBNA1 inhibition (PubChem AID-2381). Finally, we supported assessments of pocket druggability by performing induced fit docking and molecular dynamics simulations paired with binding affinity predictions by Molecular Mechanics Generalized Born Surface Area calculations for a number of hits belonging to druggable binding sites. Our results establish EBNA1 as a target for drug discovery, and provide the computational evidence that active AID-2381 hits disrupt EBNA1:DNA binding upon interacting at individual sites. Lastly, structural properties of top scoring hits are proposed to support the rational design of the next generation of EBNA1 inhibitors.
Keywords: Computational approaches to pocket finding; EBNA1 “druggability” assessment; Molecular docking; Structure-based drug design.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

