Non-coding Double-stranded RNA and Antimicrobial Peptide LL-37 Induce Growth Factor Expression from Keratinocytes and Endothelial Cells
- PMID: 27048655
- PMCID: PMC4882433
- DOI: 10.1074/jbc.M116.725317
Non-coding Double-stranded RNA and Antimicrobial Peptide LL-37 Induce Growth Factor Expression from Keratinocytes and Endothelial Cells
Abstract
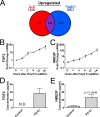
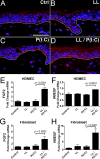
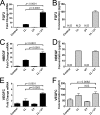
A critical function for skin is that when damaged it must simultaneously identify the nature of the injury, repair barrier function, and limit the intrusion of pathogenic organisms. These needs are carried out through the detection of damage-associated molecular patterns (DAMPs) and a response that includes secretion of cytokines, chemokines, growth factors, and antimicrobial peptides (AMPs). In this study, we analyzed how non-coding double-stranded RNA (dsRNAs) act as a DAMP in the skin and how the human cathelicidin AMP LL-37 might influence growth factor production in response to this DAMP. dsRNA alone significantly increased the expression of multiple growth factors in keratinocytes, endothelial cells, and fibroblasts. Furthermore, RNA sequencing transcriptome analysis found that multiple growth factors increase when cells are exposed to both LL-37 and dsRNA, a condition that mimics normal wounding. Quantitative PCR and/or ELISA validated that growth factors expressed by keratinocytes in these conditions included, but were not limited to, basic fibroblast growth factor (FGF2), heparin-binding EGF-like growth factor (HBEGF), vascular endothelial growth factor C (VEGFC), betacellulin (BTC), EGF, epiregulin (EREG), and other members of the transforming growth factor β superfamily. These results identify a novel role for DAMPs and AMPs in the stimulation of repair and highlight the complex interactions involved in the wound environment.
Keywords: antimicrobial peptide (AMP); betacellulin; cathelicidin; double-stranded RNA (dsRNA); fibroblast growth factor (FGF); insulin-like growth factor (IGF); keratinocyte; vascular endothelial growth factor (VEGF).
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
LL-37 regulates the overexpression of vascular endothelial growth factor (VEGF) and c-IAP-2 in human keratinocytes.Int J Dermatol. 2008 May;47(5):457-62. doi: 10.1111/j.1365-4632.2008.03340.x. Int J Dermatol. 2008. PMID: 18412861
-
Cathelicidin antimicrobial peptide LL-37 augments interferon-β expression and antiviral activity induced by double-stranded RNA in keratinocytes.Br J Dermatol. 2014 Sep;171(3):492-8. doi: 10.1111/bjd.12942. Epub 2014 Aug 5. Br J Dermatol. 2014. PMID: 24601852
-
Human antimicrobial peptide LL-37 modulates proinflammatory responses induced by cytokine milieus and double-stranded RNA in human keratinocytes.Biochem Biophys Res Commun. 2013 Apr 19;433(4):532-7. doi: 10.1016/j.bbrc.2013.03.024. Epub 2013 Mar 20. Biochem Biophys Res Commun. 2013. PMID: 23524263
-
Psoriasis and Antimicrobial Peptides.Int J Mol Sci. 2020 Sep 16;21(18):6791. doi: 10.3390/ijms21186791. Int J Mol Sci. 2020. PMID: 32947991 Free PMC article. Review.
-
AMP-IBP5: A Multifunctional Antimicrobial Peptide for Advanced Wound Healing and Inflammatory Skin Disorders.J Funct Biomater. 2025 May 12;16(5):174. doi: 10.3390/jfb16050174. J Funct Biomater. 2025. PMID: 40422838 Free PMC article. Review.
Cited by
-
Expression and Function of Host Defense Peptides at Inflammation Sites.Int J Mol Sci. 2019 Dec 22;21(1):104. doi: 10.3390/ijms21010104. Int J Mol Sci. 2019. PMID: 31877866 Free PMC article. Review.
-
The parathyroid hormone family member TIP39 interacts with sarco/endoplasmic reticulum Ca2+ - ATPase activity by influencing calcium homoeostasis.Exp Dermatol. 2017 Sep;26(9):792-797. doi: 10.1111/exd.13294. Epub 2017 Apr 21. Exp Dermatol. 2017. PMID: 28094886 Free PMC article.
-
naRNA-LL37 composite DAMPs define sterile NETs as self-propagating drivers of inflammation.EMBO Rep. 2024 Jul;25(7):2914-2949. doi: 10.1038/s44319-024-00150-5. Epub 2024 May 23. EMBO Rep. 2024. PMID: 38783164 Free PMC article.
-
Activation of Parathyroid Hormone 2 Receptor Induces Decorin Expression and Promotes Wound Repair.J Invest Dermatol. 2017 Aug;137(8):1774-1783. doi: 10.1016/j.jid.2017.03.034. Epub 2017 Apr 26. J Invest Dermatol. 2017. PMID: 28454729 Free PMC article.
-
Modulation of toll-like receptor signaling by antimicrobial peptides.Semin Cell Dev Biol. 2019 Apr;88:173-184. doi: 10.1016/j.semcdb.2018.02.002. Epub 2018 Feb 12. Semin Cell Dev Biol. 2019. PMID: 29432957 Free PMC article. Review.
References
-
- Fulton C., Anderson G. M., Zasloff M., Bull R., and Quinn A. G. (1997) Expression of natural peptide antibiotics in human skin. Lancet 350, 1750–1751 - PubMed
-
- Harder J., Bartels J., Christophers E., and Schröder J. M. (1997) A peptide antibiotic from human skin. Nature 387, 861. - PubMed
-
- Harder J., Bartels J., Christophers E., and Schroder J. M. (2001) Isolation and characterization of human β-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276, 5707–5713 - PubMed
-
- Frohm M., Agerberth B., Ahangari G., Stâhle-Bäckdahl M., Lidén S., Wigzell H., and Gudmundsson G. H. (1997) The expression of the gene coding for the antibacterial peptide LL-37 is induced in human keratinocytes during inflammatory disorders. J. Biol. Chem. 272, 15258–15263 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

