LGR5 expression is controled by IKKα in basal cell carcinoma through activating STAT3 signaling pathway
- PMID: 27049829
- PMCID: PMC5053649
- DOI: 10.18632/oncotarget.8465
LGR5 expression is controled by IKKα in basal cell carcinoma through activating STAT3 signaling pathway
Abstract
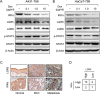
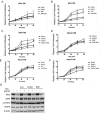
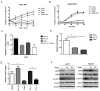
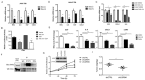
Basal cell carcinomas (BCC) of the skin are the most common of human cancers. The noncanonical NF-κB pathway is dependent on IKKα. However, the role of IKKα in BCC has not been elucidated. We show here that IKKα is expressed in the nucleus in BCC and non-malignant diseases. Nuclear IKKα could directly bind to the promoters of inflammation factors and LGR5, a stem cell marker, in turn, upregulating LGR5 expression through activation of STAT3 signaling pathway during cancer progression. Activation of STAT3 signaling pathway contributes LGR5 expression in dependent of IKKα after the interplay between STAT3 and IKKα. Meanwhile knockdown of IKKα inhibits tumor growth and transition of epithelial stage to mescheme stage. Taken together, we demonstrate that IKKα functions as a bone fide chromatin regulator in BCC, whose promoted expression contributes to oncogenic transformation via promoting expression stemness- and inflammatory- related genes. Our finding reveals a novel viewpoint for how IKKα may involve in BCCs tumor progression in the inflammatory microenvironment.
Keywords: BCC; EGF; IKKα; LGR5; STAT3.
Conflict of interest statement
The authors declare no conflict of interest. This manuscript has been read and approved by all the authors, and not submitted or under consider for publication elsewhere.
Figures


Similar articles
-
A role for transcription factor STAT3 signaling in oncogene smoothened-driven carcinogenesis.J Biol Chem. 2012 Nov 2;287(45):38356-66. doi: 10.1074/jbc.M112.377382. Epub 2012 Sep 19. J Biol Chem. 2012. PMID: 22992748 Free PMC article.
-
Targeting Notch1 and IKKα Enhanced NF-κB Activation in CD133+ Skin Cancer Stem Cells.Mol Cancer Ther. 2018 Sep;17(9):2034-2048. doi: 10.1158/1535-7163.MCT-17-0421. Epub 2018 Jun 29. Mol Cancer Ther. 2018. PMID: 29959199 Free PMC article.
-
Nuclear versus cytoplasmic IKKα signaling in keratinocytes leads to opposite skin phenotypes and inflammatory responses, and a different predisposition to cancer.Oncogene. 2025 Feb;44(3):165-178. doi: 10.1038/s41388-024-03203-0. Epub 2024 Nov 7. Oncogene. 2025. PMID: 39511409 Free PMC article.
-
Role of IKKα in skin squamous cell carcinomas.Future Oncol. 2011 Jan;7(1):123-34. doi: 10.2217/fon.10.166. Future Oncol. 2011. PMID: 21174543 Free PMC article. Review.
-
Understanding the Molecular Genetics of Basal Cell Carcinoma.Int J Mol Sci. 2017 Nov 22;18(11):2485. doi: 10.3390/ijms18112485. Int J Mol Sci. 2017. PMID: 29165358 Free PMC article. Review.
Cited by
-
DDX5 Silencing Suppresses the Migration of Basal cell Carcinoma Cells by Downregulating JAK2/STAT3 Pathway.Technol Cancer Res Treat. 2019 Jan-Dec;18:1533033819892258. doi: 10.1177/1533033819892258. Technol Cancer Res Treat. 2019. PMID: 31870221 Free PMC article.
-
The value of detecting immunoglobulin gene rearrangements in the diagnosis of B-cell lymphoma.Oncotarget. 2017 Aug 18;8(44):77009-77019. doi: 10.18632/oncotarget.20330. eCollection 2017 Sep 29. Oncotarget. 2017. PMID: 29100365 Free PMC article.
-
IL-22 initiates an IL-18-dependent epithelial response circuit to enforce intestinal host defence.Nat Commun. 2022 Feb 15;13(1):874. doi: 10.1038/s41467-022-28478-3. Nat Commun. 2022. PMID: 35169117 Free PMC article.
-
NF-κB Members Left Home: NF-κB-Independent Roles in Cancer.Biomedicines. 2017 May 25;5(2):26. doi: 10.3390/biomedicines5020026. Biomedicines. 2017. PMID: 28587092 Free PMC article. Review.
-
NFκB signalling in colorectal cancer: Examining the central dogma of IKKα and IKKβ signalling.Heliyon. 2024 Jun 12;10(12):e32904. doi: 10.1016/j.heliyon.2024.e32904. eCollection 2024 Jun 30. Heliyon. 2024. PMID: 38975078 Free PMC article. Review.
References
-
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. - PubMed
-
- Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, Toftgard R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–1299. - PubMed
-
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. - PubMed
-
- Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nature reviews Molecular cell biology. 2007;8:49–62. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

