Molecular crosstalk between tumour and brain parenchyma instructs histopathological features in glioblastoma
- PMID: 27049916
- PMCID: PMC5077988
- DOI: 10.18632/oncotarget.7454
Molecular crosstalk between tumour and brain parenchyma instructs histopathological features in glioblastoma
Abstract
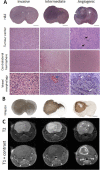
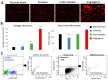
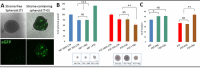
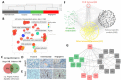
The histopathological and molecular heterogeneity of glioblastomas represents a major obstacle for effective therapies. Glioblastomas do not develop autonomously, but evolve in a unique environment that adapts to the growing tumour mass and contributes to the malignancy of these neoplasms. Here, we show that patient-derived glioblastoma xenografts generated in the mouse brain from organotypic spheroids reproducibly give rise to three different histological phenotypes: (i) a highly invasive phenotype with an apparent normal brain vasculature, (ii) a highly angiogenic phenotype displaying microvascular proliferation and necrosis and (iii) an intermediate phenotype combining features of invasion and vessel abnormalities. These phenotypic differences were visible during early phases of tumour development suggesting an early instructive role of tumour cells on the brain parenchyma. Conversely, we found that tumour-instructed stromal cells differentially influenced tumour cell proliferation and migration in vitro, indicating a reciprocal crosstalk between neoplastic and non-neoplastic cells. We did not detect any transdifferentiation of tumour cells into endothelial cells. Cell type-specific transcriptomic analysis of tumour and endothelial cells revealed a strong phenotype-specific molecular conversion between the two cell types, suggesting co-evolution of tumour and endothelial cells. Integrative bioinformatic analysis confirmed the reciprocal crosstalk between tumour and microenvironment and suggested a key role for TGFβ1 and extracellular matrix proteins as major interaction modules that shape glioblastoma progression. These data provide novel insight into tumour-host interactions and identify novel stroma-specific targets that may play a role in combinatorial treatment strategies against glioblastoma.
Keywords: angiogenesis; endothelial cells; glioblastoma; patient-derived xenograft; tumour microenvironment.
Conflict of interest statement
Authors have no conflict of interest to disclose
Figures


References
-
- De Witt Hamer PC, Van Tilborg AA, Eijk PP, Sminia P, Troost D, Van Noorden CJ, Ylstra B, Leenstra S. The genomic profile of human malignant glioma is altered early in primary cell culture and preserved in spheroids. Oncogene. 2008;27:2091–2096. - PubMed
-
- Sakariassen PO, Prestegarden L, Wang J, Skaftnesmo KO, Mahesparan R, Molthoff C, Sminia P, Sundlisaeter E, Misra A, Tysnes BB, Chekenya M, Peters H, Lende G, Kalland KH, Oyan AM, Petersen K, et al. Angiogenesis-independent tumor growth mediated by stem-like cancer cells. Proc Natl Acad Sci U S A. 2006;103:16466–16471. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

