A Querying Method over RDF-ized Health Level Seven v2.5 Messages Using Life Science Knowledge Resources
- PMID: 27050304
- PMCID: PMC4837294
- DOI: 10.2196/medinform.5275
A Querying Method over RDF-ized Health Level Seven v2.5 Messages Using Life Science Knowledge Resources
Abstract
Background: Health level seven version 2.5 (HL7 v2.5) is a widespread messaging standard for information exchange between clinical information systems. By applying Semantic Web technologies for handling HL7 v2.5 messages, it is possible to integrate large-scale clinical data with life science knowledge resources.
Objective: Showing feasibility of a querying method over large-scale resource description framework (RDF)-ized HL7 v2.5 messages using publicly available drug databases.
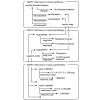
Methods: We developed a method to convert HL7 v2.5 messages into the RDF. We also converted five kinds of drug databases into RDF and provided explicit links between the corresponding items among them. With those linked drug data, we then developed a method for query expansion to search the clinical data using semantic information on drug classes along with four types of temporal patterns. For evaluation purpose, medication orders and laboratory test results for a 3-year period at the University of Tokyo Hospital were used, and the query execution times were measured.
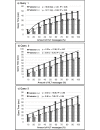
Results: Approximately 650 million RDF triples for medication orders and 790 million RDF triples for laboratory test results were converted. Taking three types of query in use cases for detecting adverse events of drugs as an example, we confirmed these queries were represented in SPARQL Protocol and RDF Query Language (SPARQL) using our methods and comparison with conventional query expressions were performed. The measurement results confirm that the query time is feasible and increases logarithmically or linearly with the amount of data and without diverging.
Conclusions: The proposed methods enabled query expressions that separate knowledge resources and clinical data, thereby suggesting the feasibility for improving the usability of clinical data by enhancing the knowledge resources. We also demonstrate that when HL7 v2.5 messages are automatically converted into RDF, searches are still possible through SPARQL without modifying the structure. As such, the proposed method benefits not only our hospitals, but also numerous hospitals that handle HL7 v2.5 messages. Our approach highlights a potential of large-scale data federation techniques to retrieve clinical information, which could be applied as applications of clinical intelligence to improve clinical practices, such as adverse drug event monitoring and cohort selection for a clinical study as well as discovering new knowledge from clinical information.
Keywords: Semantic Web; electronic health records; health level seven; information storage and retrieval; linked open data.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures






References
-
- Badawi O, Brennan T, Celi LA, Feng M, Ghassemi M, Ippolito A, Johnson A, Mark RG, Mayaud L, Moody G, Moses C, Naumann T, Pimentel M, Pollard TJ, Santos M, Stone DJ, Zimolzak A, MIT Critical Data Conference 2014 Organizing Committee Making big data useful for health care: a summary of the inaugural mit critical data conference. JMIR Med Inform. 2014;2:e22. doi: 10.2196/medinform.3447. http://medinform.jmir.org/2014/2/e22/ v2i2e22 - DOI - PMC - PubMed
-
- Moseley ET, Hsu DJ, Stone DJ, Celi LA. Beyond open big data: addressing unreliable research. J Med Internet Res. 2014;16:e259. doi: 10.2196/jmir.3871. http://www.jmir.org/2014/11/e259/ v16i11e259 - DOI - PMC - PubMed
-
- Bizer C, Heath T, Berners-Lee T. Linked data - the story so far. International Journal on Semantic Web and Information Systems. 2009;5:1–22. doi: 10.4018/jswis.2009081901. - DOI
-
- Belleau F, Nolin M, Tourigny N, Rigault P, Morissette J. Bio2RDF: towards a mashup to build bioinformatics knowledge systems. J Biomed Inform. 2008;41:706–716. doi: 10.1016/j.jbi.2008.03.004. http://linkinghub.elsevier.com/retrieve/pii/S1532-0464(08)00041-5 S1532-0464(08)00041-5 - DOI - PubMed
-
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. http://nar.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=10592173 gkd027 - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

