Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs
- PMID: 27050392
- PMCID: PMC4823868
- DOI: 10.1038/ncomms11215
Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs
Abstract
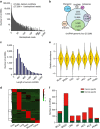
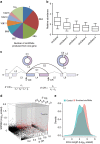
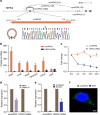
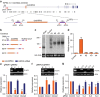
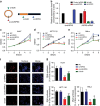
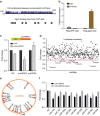
Circular RNAs (circRNAs) represent a class of widespread and diverse endogenous RNAs that may regulate gene expression in eukaryotes. However, the regulation and function of human circRNAs remain largely unknown. Here we generate ribosomal-depleted RNA sequencing data from six normal tissues and seven cancers, and detect at least 27,000 circRNA candidates. Many of these circRNAs are differently expressed between the normal and cancerous tissues. We further characterize one abundant circRNA derived from Exon2 of the HIPK3 gene, termed circHIPK3. The silencing of circHIPK3 but not HIPK3 mRNA significantly inhibits human cell growth. Via a luciferase screening assay, circHIPK3 is observed to sponge to 9 miRNAs with 18 potential binding sites. Specifically, we show that circHIPK3 directly binds to miR-124 and inhibits miR-124 activity. Our results provide evidence that circular RNA produced from precursor mRNA may have a regulatory role in human cells.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

