A lysosome-centered view of nutrient homeostasis
- PMID: 27050453
- PMCID: PMC4836021
- DOI: 10.1080/15548627.2016.1147671
A lysosome-centered view of nutrient homeostasis
Abstract
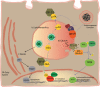
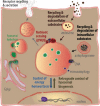
Lysosomes are highly acidic cellular organelles traditionally viewed as sacs of enzymes involved in digesting extracellular or intracellular macromolecules for the regeneration of basic building blocks, cellular housekeeping, or pathogen degradation. Bound by a single lipid bilayer, lysosomes receive their substrates by fusing with endosomes or autophagosomes, or through specialized translocation mechanisms such as chaperone-mediated autophagy or microautophagy. Lysosomes degrade their substrates using up to 60 different soluble hydrolases and release their products either to the cytosol through poorly defined exporting and efflux mechanisms or to the extracellular space by fusing with the plasma membrane. However, it is becoming evident that the role of the lysosome in nutrient homeostasis goes beyond the disposal of waste or the recycling of building blocks. The lysosome is emerging as a signaling hub that can integrate and relay external and internal nutritional information to promote cellular and organismal homeostasis, as well as a major contributor to the processing of energy-dense molecules like glycogen and triglycerides. Here we describe the current knowledge of the nutrient signaling pathways governing lysosomal function, the role of the lysosome in nutrient mobilization, and how lysosomes signal other organelles, distant tissues, and even themselves to ensure energy homeostasis in spite of fluctuations in energy intake. At the same time, we highlight the value of genomics approaches to the past and future discoveries of how the lysosome simultaneously executes and controls cellular homeostasis.
Keywords: C elegans; amino acid; homeostasis; human; hydrolase; lipid; lysosome; metabolism; nutrient; sensor; yeast.
Figures

References
-
- Sleat DE, Della Valle MC, Zheng H, Moore DF, Lobel P. The mannose 6-phosphate glycoprotein proteome. J Proteome Res 2008; 7:3010-21; PMID:18507433; http://dx.doi.org/10.1021/pr800135v - DOI - PMC - PubMed
-
- Eskelinen EL, Tanaka Y, Saftig P. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol 2003; 13:137-45; PMID:12628346; http://dx.doi.org/10.1016/S0962-8924(03)00005-9 - DOI - PubMed
-
- Bainton DF. The Discovery of Lysosomes. J Cell Biol 1981; 91:S66-S76; http://dx.doi.org/10.1083/jcb.91.3.66s - DOI - PMC - PubMed
-
- Okamoto K. Organellophagy: eliminating cellular building blocks via selective autophagy. J Cell Biol 2014; 205:435-45; PMID:24862571; http://dx.doi.org/10.1083/jcb.201402054 - DOI - PMC - PubMed
-
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 2011; 12:21-35; PMID:21157483; http://dx.doi.org/10.1038/nrm3025 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
