Design and computational analysis of single-cell RNA-sequencing experiments
- PMID: 27052890
- PMCID: PMC4823857
- DOI: 10.1186/s13059-016-0927-y
Design and computational analysis of single-cell RNA-sequencing experiments
Abstract
Single-cell RNA-sequencing (scRNA-seq) has emerged as a revolutionary tool that allows us to address scientific questions that eluded examination just a few years ago. With the advantages of scRNA-seq come computational challenges that are just beginning to be addressed. In this article, we highlight the computational methods available for the design and analysis of scRNA-seq experiments, their advantages and disadvantages in various settings, the open questions for which novel methods are needed, and expected future developments in this exciting area.
Figures
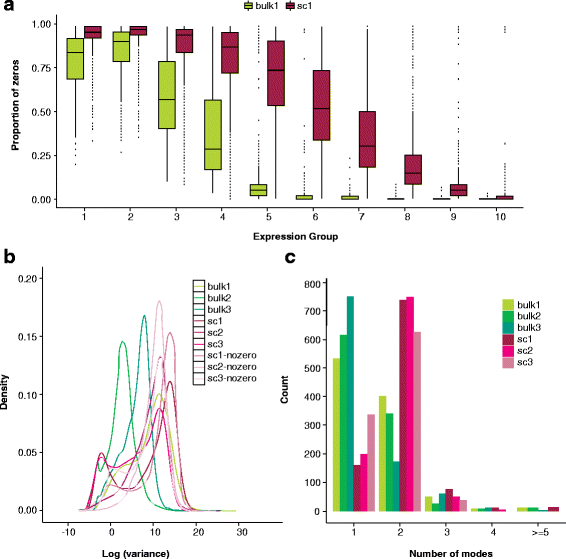
References
-
- Hicks SC, Teng M, Irizarry RA. On the widespread and critical impact of systematic bias and batch effects in single-cell RNA-Seq data. bioRxiv. 2015. doi: http://dx.doi.org/10.1101/025528. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

