A dual drug regimen synergistically blocks human parainfluenza virus infection
- PMID: 27053240
- PMCID: PMC4823791
- DOI: 10.1038/srep24138
A dual drug regimen synergistically blocks human parainfluenza virus infection
Abstract
Human parainfluenza type-3 virus (hPIV-3) is one of the principal aetiological agents of acute respiratory illness in infants worldwide and also shows high disease severity in the elderly and immunocompromised, but neither therapies nor vaccines are available to treat or prevent infection, respectively. Using a multidisciplinary approach we report herein that the approved drug suramin acts as a non-competitive in vitro inhibitor of the hPIV-3 haemagglutinin-neuraminidase (HN). Furthermore, the drug inhibits viral replication in mammalian epithelial cells with an IC50 of 30 μM, when applied post-adsorption. Significantly, we show in cell-based drug-combination studies using virus infection blockade assays, that suramin acts synergistically with the anti-influenza virus drug zanamivir. Our data suggests that lower concentrations of both drugs can be used to yield high levels of inhibition. Finally, using NMR spectroscopy and in silico docking simulations we confirmed that suramin binds HN simultaneously with zanamivir. This binding event occurs most likely in the vicinity of the protein primary binding site, resulting in an enhancement of the inhibitory potential of the N-acetylneuraminic acid-based inhibitor. This study offers a potentially exciting avenue for the treatment of parainfluenza infection by a combinatorial repurposing approach of well-established approved drugs.
Figures

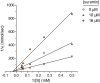
 as a function of
as a function of  according to Equation (1), and is representative of 3 independent experiments. The straight lines are linear regressions calculated for each concentration of inhibitor.
according to Equation (1), and is representative of 3 independent experiments. The straight lines are linear regressions calculated for each concentration of inhibitor.
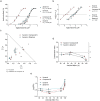


References
-
- Hall C. B. Respiratory syncytial virus and parainfluenza virus. N. Engl. J. Med. 344, 1917–1928 (2001). - PubMed
-
- Cunha B. A., Corbett M. & Mickail N. Human parainfluenza virus type 3 (HPIV 3) viral community-acquired pneumonia (CAP) mimicking swine influenza (H1N1) during the swine flu pandemic. Heart Lung 40, 76–80 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

