Mutational History of a Human Cell Lineage from Somatic to Induced Pluripotent Stem Cells
- PMID: 27054363
- PMCID: PMC4824386
- DOI: 10.1371/journal.pgen.1005932
Mutational History of a Human Cell Lineage from Somatic to Induced Pluripotent Stem Cells
Abstract
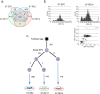
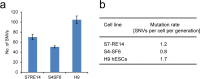
The accuracy of replicating the genetic code is fundamental. DNA repair mechanisms protect the fidelity of the genome ensuring a low error rate between generations. This sustains the similarity of individuals whilst providing a repertoire of variants for evolution. The mutation rate in the human genome has recently been measured to be 50-70 de novo single nucleotide variants (SNVs) between generations. During development mutations accumulate in somatic cells so that an organism is a mosaic. However, variation within a tissue and between tissues has not been analysed. By reprogramming somatic cells into induced pluripotent stem cells (iPSCs), their genomes and the associated mutational history are captured. By sequencing the genomes of polyclonal and monoclonal somatic cells and derived iPSCs we have determined the mutation rates and show how the patterns change from a somatic lineage in vivo through to iPSCs. Somatic cells have a mutation rate of 14 SNVs per cell per generation while iPSCs exhibited a ten-fold lower rate. Analyses of mutational signatures suggested that deamination of methylated cytosine may be the major mutagenic source in vivo, whilst oxidative DNA damage becomes dominant in vitro. Our results provide insights for better understanding of mutational processes and lineage relationships between human somatic cells. Furthermore it provides a foundation for interpretation of elevated mutation rates and patterns in cancer.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

