Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer
- PMID: 27055085
- PMCID: PMC4982795
- DOI: 10.1001/jamaoncol.2016.0173
Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer
Erratum in
-
Error in Funding Statement.JAMA Oncol. 2016 Aug 1;2(8):1099. doi: 10.1001/jamaoncol.2016.1752. JAMA Oncol. 2016. PMID: 27244704 No abstract available.
Abstract
Importance: Plasma genotyping of cell-free DNA has the potential to allow for rapid noninvasive genotyping while avoiding the inherent shortcomings of tissue genotyping and repeat biopsies.
Objective: To prospectively validate plasma droplet digital PCR (ddPCR) for the rapid detection of common epidermal growth factor receptor (EGFR) and KRAS mutations, as well as the EGFR T790M acquired resistance mutation.
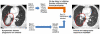
Design, setting, and participants: Patients with advanced nonsquamous non-small-cell lung cancer (NSCLC) who either (1) had a new diagnosis and were planned for initial therapy or (2) had developed acquired resistance to an EGFR kinase inhibitor and were planned for rebiopsy underwent initial blood sampling and immediate plasma ddPCR for EGFR exon 19 del, L858R, T790M, and/or KRAS G12X between July 3, 2014, and June 30, 2015, at a National Cancer Institute-designated comprehensive cancer center. All patients underwent biopsy for tissue genotyping, which was used as the reference standard for comparison; rebiopsy was required for patients with acquired resistance to EGFR kinase inhibitors. Test turnaround time (TAT) was measured in business days from blood sampling until test reporting.
Main outcomes and measures: Plasma ddPCR assay sensitivity, specificity, and TAT.
Results: Of 180 patients with advanced NSCLC (62% female; median [range] age, 62 [37-93] years), 120 cases were newly diagnosed; 60 had acquired resistance. Tumor genotype included 80 EGFR exon 19/L858R mutants, 35 EGFR T790M, and 25 KRAS G12X mutants. Median (range) TAT for plasma ddPCR was 3 (1-7) days. Tissue genotyping median (range) TAT was 12 (1-54) days for patients with newly diagnosed NSCLC and 27 (1-146) days for patients with acquired resistance. Plasma ddPCR exhibited a positive predictive value of 100% (95% CI, 91%-100%) for EGFR 19 del, 100% (95% CI, 85%-100%) for L858R, and 100% (95% CI, 79%-100%) for KRAS, but lower for T790M at 79% (95% CI, 62%-91%). The sensitivity of plasma ddPCR was 82% (95% CI, 69%-91%) for EGFR 19 del, 74% (95% CI, 55%-88%) for L858R, and 77% (95% CI, 60%-90%) for T790M, but lower for KRAS at 64% (95% CI, 43%-82%). Sensitivity for EGFR or KRAS was higher in patients with multiple metastatic sites and those with hepatic or bone metastases, specifically.
Conclusions and relevance: Plasma ddPCR detected EGFR and KRAS mutations rapidly with the high specificity needed to select therapy and avoid repeat biopsies. This assay may also detect EGFR T790M missed by tissue genotyping due to tumor heterogeneity in resistant disease.
Figures
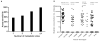

Comment in
-
Lung cancer: Using ctDNA to track EGFR and KRAS mutations in advanced-stage disease.Nat Rev Clin Oncol. 2016 Jul;13(7):401-2. doi: 10.1038/nrclinonc.2016.83. Epub 2016 Jun 1. Nat Rev Clin Oncol. 2016. PMID: 27245284 No abstract available.
-
Early Intervention in Lung Cancers With Rapid Plasma Genotyping for EGFR and KRAS Mutations.JAMA Oncol. 2016 Aug 1;2(8):1096. doi: 10.1001/jamaoncol.2016.2161. JAMA Oncol. 2016. PMID: 27388195 No abstract available.
References
-
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

