Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions
- PMID: 27056827
- PMCID: PMC4823848
- DOI: 10.1186/s13073-016-0290-3
Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions
Abstract
Background: Colorectal cancer (CRC) is the second leading cause of death among cancers in the United States. Although individuals diagnosed early have a greater than 90% chance of survival, more than one-third of individuals do not adhere to screening recommendations partly because the standard diagnostics, colonoscopy and sigmoidoscopy, are expensive and invasive. Thus, there is a great need to improve the sensitivity of non-invasive tests to detect early stage cancers and adenomas. Numerous studies have identified shifts in the composition of the gut microbiota associated with the progression of CRC, suggesting that the gut microbiota may represent a reservoir of biomarkers that would complement existing non-invasive methods such as the widely used fecal immunochemical test (FIT).
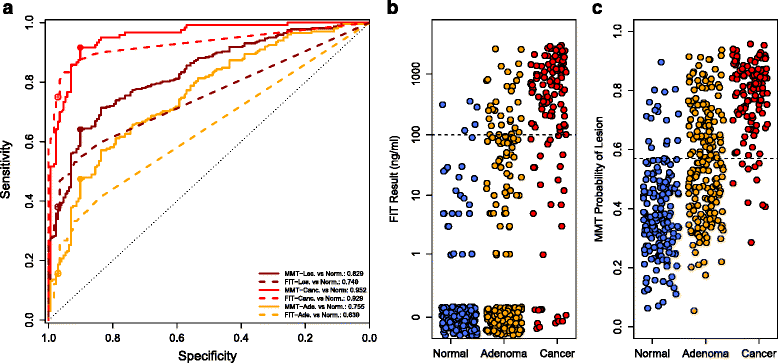
Methods: We sequenced the 16S rRNA genes from the stool samples of 490 patients. We used the relative abundances of the bacterial populations within each sample to develop a random forest classification model that detects colonic lesions using the relative abundance of gut microbiota and the concentration of hemoglobin in stool.
Results: The microbiota-based random forest model detected 91.7% of cancers and 45.5% of adenomas while FIT alone detected 75.0% and 15.7%, respectively. Of the colonic lesions missed by FIT, the model detected 70.0% of cancers and 37.7% of adenomas. We confirmed known associations of Porphyromonas assaccharolytica, Peptostreptococcus stomatis, Parvimonas micra, and Fusobacterium nucleatum with CRC. Yet, we found that the loss of potentially beneficial organisms, such as members of the Lachnospiraceae, was more predictive for identifying patients with adenomas when used in combination with FIT.
Conclusions: These findings demonstrate the potential for microbiota analysis to complement existing screening methods to improve detection of colonic lesions.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

