Mutations and altered expression of SERPINF1 in patients with familial otosclerosis
- PMID: 27056980
- PMCID: PMC5181625
- DOI: 10.1093/hmg/ddw106
Mutations and altered expression of SERPINF1 in patients with familial otosclerosis
Abstract
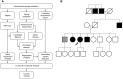
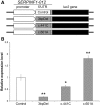
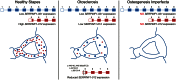
Otosclerosis is a relatively common heterogenous condition, characterized by abnormal bone remodelling in the otic capsule leading to fixation of the stapedial footplate and an associated conductive hearing loss. Although familial linkage and candidate gene association studies have been performed in recent years, little progress has been made in identifying disease-causing genes. Here, we used whole-exome sequencing in four families exhibiting dominantly inherited otosclerosis to identify 23 candidate variants (reduced to 9 after segregation analysis) for further investigation in a secondary cohort of 84 familial cases. Multiple mutations were found in the SERPINF1 (Serpin Peptidase Inhibitor, Clade F) gene which encodes PEDF (pigment epithelium-derived factor), a potent inhibitor of angiogenesis and known regulator of bone density. Six rare heterozygous SERPINF1 variants were found in seven patients in our familial otosclerosis cohort; three are missense mutations predicted to be deleterious to protein function. The other three variants are all located in the 5'-untranslated region (UTR) of an alternative spliced transcript SERPINF1-012 RNA-seq analysis demonstrated that this is the major SERPINF1 transcript in human stapes bone. Analysis of stapes from two patients with the 5'-UTR mutations showed that they had reduced expression of SERPINF1-012 All three 5'-UTR mutations are predicted to occur within transcription factor binding sites and reporter gene assays confirmed that they affect gene expression levels. Furthermore, RT-qPCR analysis of stapes bone cDNA showed that SERPINF1-012 expression is reduced in otosclerosis patients with and without SERPINF1 mutations, suggesting that it may be a common pathogenic pathway in the disease.
© The Author 2016. Published by Oxford University Press.
Figures

References
-
- Nager G.T. (1969) Histopathology of otosclerosis. Arch. Otolaryngol., 89, 341–363. - PubMed
-
- Declau F., Van Spaendonck M., Timmermans J.P., Michaels L., Liang J., Qiu J.P., Van De Heyning P. (2007) Prevalence of histologic otosclerosis: An unbiased temporal bone study in caucasians. Adv. Otorhinolaryngol., 65, 6–16. - PubMed
-
- Emmett J.R. (1993) Physical examination and clinical evaluation of the patient with otosclerosis. Otolaryngol. Clin. North Am., 26, 353–357. - PubMed
-
- Ealy M., Smith R.J.H. (2011) Otosclerosis. Adv. Otorhinolaryngol., 70, 122–129. - PubMed
-
- Gordon M.A. (1989) The genetics of otosclerosis: a review. Am. J. Otol., 10, 426–438. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

