Mutation-Specific Phenotypes in hiPSC-Derived Cardiomyocytes Carrying Either Myosin-Binding Protein C Or α-Tropomyosin Mutation for Hypertrophic Cardiomyopathy
- PMID: 27057166
- PMCID: PMC4707351
- DOI: 10.1155/2016/1684792
Mutation-Specific Phenotypes in hiPSC-Derived Cardiomyocytes Carrying Either Myosin-Binding Protein C Or α-Tropomyosin Mutation for Hypertrophic Cardiomyopathy
Abstract
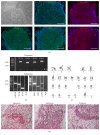
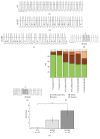
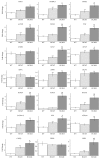
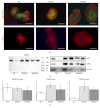
Hypertrophic cardiomyopathy (HCM) is a genetic cardiac disease, which affects the structure of heart muscle tissue. The clinical symptoms include arrhythmias, progressive heart failure, and even sudden cardiac death but the mutation carrier can also be totally asymptomatic. To date, over 1400 mutations have been linked to HCM, mostly in genes encoding for sarcomeric proteins. However, the pathophysiological mechanisms of the disease are still largely unknown. Two founder mutations for HCM in Finland are located in myosin-binding protein C (MYBPC3-Gln1061X) and α-tropomyosin (TPM1-Asp175Asn) genes. We studied the properties of HCM cardiomyocytes (CMs) derived from patient-specific human induced pluripotent stem cells (hiPSCs) carrying either MYBPC3-Gln1061X or TPM1-Asp175Asn mutation. Both types of HCM-CMs displayed pathological phenotype of HCM but, more importantly, we found differences between CMs carrying either MYBPC3-Gln1061X or TPM1-Asp175Asn gene mutation in their cellular size, Ca(2+) handling, and electrophysiological properties, as well as their gene expression profiles. These findings suggest that even though the clinical phenotypes of the patients carrying either MYBPC3-Gln1061X or TPM1-Asp175Asn gene mutation are similar, the genetic background as well as the functional properties on the cellular level might be different, indicating that the pathophysiological mechanisms behind the two mutations would be divergent as well.
Figures

Similar articles
-
Divergent effects of adrenaline in human induced pluripotent stem cell-derived cardiomyocytes obtained from hypertrophic cardiomyopathy.Dis Model Mech. 2018 Feb 26;11(2):dmm032896. doi: 10.1242/dmm.032896. Dis Model Mech. 2018. PMID: 29361520 Free PMC article.
-
Genetics of hypertrophic cardiomyopathy in eastern Finland: few founder mutations with benign or intermediary phenotypes.Ann Med. 2004;36(1):23-32. doi: 10.1080/07853890310017161. Ann Med. 2004. PMID: 15000344 Review.
-
Mutations in the cardiac myosin-binding protein C gene are the predominant cause of familial hypertrophic cardiomyopathy in eastern Finland.J Mol Med (Berl). 2002 Jul;80(7):412-22. doi: 10.1007/s00109-002-0323-9. Epub 2002 Apr 11. J Mol Med (Berl). 2002. PMID: 12110947
-
Two founder mutations in the alpha-tropomyosin and the cardiac myosin-binding protein C genes are common causes of hypertrophic cardiomyopathy in the Finnish population.Ann Med. 2013 Feb;45(1):85-90. doi: 10.3109/07853890.2012.671534. Epub 2012 Apr 2. Ann Med. 2013. PMID: 22462493
-
Clinical outcomes associated with sarcomere mutations in hypertrophic cardiomyopathy: a meta-analysis on 7675 individuals.Clin Res Cardiol. 2018 Jan;107(1):30-41. doi: 10.1007/s00392-017-1155-5. Epub 2017 Aug 24. Clin Res Cardiol. 2018. PMID: 28840316 Review.
Cited by
-
Divergent effects of adrenaline in human induced pluripotent stem cell-derived cardiomyocytes obtained from hypertrophic cardiomyopathy.Dis Model Mech. 2018 Feb 26;11(2):dmm032896. doi: 10.1242/dmm.032896. Dis Model Mech. 2018. PMID: 29361520 Free PMC article.
-
The Junctophilin-2 Mutation p.(Thr161Lys) Is Associated with Hypertrophic Cardiomyopathy Using Patient-Specific iPS Cardiomyocytes and Demonstrates Prolonged Action Potential and Increased Arrhythmogenicity.Biomedicines. 2023 May 27;11(6):1558. doi: 10.3390/biomedicines11061558. Biomedicines. 2023. PMID: 37371654 Free PMC article.
-
Cardiomyopathy phenotypes in human-induced pluripotent stem cell-derived cardiomyocytes-a systematic review.Pflugers Arch. 2019 May;471(5):755-768. doi: 10.1007/s00424-018-2214-0. Epub 2018 Oct 15. Pflugers Arch. 2019. PMID: 30324321 Free PMC article.
-
Experimental Models of Hypertrophic Cardiomyopathy: A Systematic Review.JACC Basic Transl Sci. 2025 Apr;10(4):511-546. doi: 10.1016/j.jacbts.2024.10.017. Epub 2025 Jan 15. JACC Basic Transl Sci. 2025. PMID: 40306862 Free PMC article. Review.
-
Disease modeling of a mutation in α-actinin 2 guides clinical therapy in hypertrophic cardiomyopathy.EMBO Mol Med. 2019 Dec;11(12):e11115. doi: 10.15252/emmm.201911115. Epub 2019 Nov 3. EMBO Mol Med. 2019. PMID: 31680489 Free PMC article.
References
-
- Jääskeläinen P., Heliö T., Aalto-Setälä K., et al. Two founder mutations in the alpha-tropomyosin and the cardiac myosin-binding protein C genes are common causes of hypertrophic cardiomyopathy in the Finnish population. Annals of Medicine. 2013;45(1):85–90. doi: 10.3109/07853890.2012.671534. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

