Medulloblastoma-associated DDX3 variant selectively alters the translational response to stress
- PMID: 27058758
- PMCID: PMC5053718
- DOI: 10.18632/oncotarget.8612
Medulloblastoma-associated DDX3 variant selectively alters the translational response to stress
Abstract
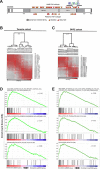
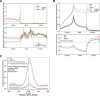
DDX3X encodes a DEAD-box family RNA helicase (DDX3) commonly mutated in medulloblastoma, a highly aggressive cerebellar tumor affecting both children and adults. Despite being implicated in several facets of RNA metabolism, the nature and scope of DDX3's interactions with RNA remain unclear. Here, we show DDX3 collaborates extensively with the translation initiation machinery through direct binding to 5'UTRs of nearly all coding RNAs, specific sites on the 18S rRNA, and multiple components of the translation initiation complex. Impairment of translation initiation is also evident in primary medulloblastomas harboring mutations in DDX3X, further highlighting DDX3's role in this process. Arsenite-induced stress shifts DDX3 binding from the 5'UTR into the coding region of mRNAs concomitant with a general reduction of translation, and both the shift of DDX3 on mRNA and decreased translation are blunted by expression of a catalytically-impaired, medulloblastoma-associated DDX3R534H variant. Furthermore, despite the global repression of translation induced by arsenite, translation is preserved on select genes involved in chromatin organization in DDX3R534H-expressing cells. Thus, DDX3 interacts extensively with RNA and ribosomal machinery to help remodel the translation landscape in response to stress, while cancer-related DDX3 variants adapt this response to selectively preserve translation.
Keywords: CLIP-seq; DDX3; DDX3X; RNA helicase; medulloblastoma.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Kool M, Jones DTW, Jäger N, Northcott PA, Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M, Milde T, Bourdeaut F, Ryzhova M, et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to Smoothened inhibition. Cancer Cell. 2014;25:393–405. doi: 10.1016/j.ccr.2014.02.004. - DOI - PMC - PubMed
-
- Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, Greulich H, Lawrence MS, Lennon NJ, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

