Mutations in the C-terminal region affect subcellular localization of crucian carp herpesvirus (CaHV) GPCR
- PMID: 27059239
- PMCID: PMC4923094
- DOI: 10.1007/s11262-016-1325-y
Mutations in the C-terminal region affect subcellular localization of crucian carp herpesvirus (CaHV) GPCR
Abstract
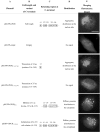
G protein-coupled receptors (GPCRs) are known as seven transmembrane domain receptors and consequently can mediate diverse biological functions via regulation of their subcellular localization. Crucian carp herpesvirus (CaHV) was recently isolated from infected fish with acute gill hemorrhage. CaHV GPCR of 349 amino acids (aa) was identified based on amino acid identity. A series of variants with truncation/deletion/substitution mutation in the C-terminal (aa 315-349) were constructed and expressed in fathead minnow (FHM) cells. The roles of three key C-terminal regions in subcellular localization of CaHV GPCR were determined. Lysine-315 (K-315) directed the aggregation of the protein preferentially at the nuclear side. Predicted N-myristoylation site (GGGWTR, aa 335-340) was responsible for punctate distribution in periplasm or throughout the cytoplasm. Predicted phosphorylation site (SSR, aa 327-329) and GGGWTR together determined the punctate distribution in cytoplasm. Detection of organelles localization by specific markers showed that the protein retaining K-315 colocalized with the Golgi apparatus. These experiments provided first evidence that different mutations of CaHV GPCR C-terminals have different affects on the subcellular localization of fish herpesvirus-encoded GPCRs. The study provided valuable information and new insights into the precise interactions between herpesvirus and fish cells, and could also provide useful targets for antiviral agents in aquaculture.
Keywords: C-terminal; Crucian carp herpesvirus (CaHV); G protein-coupled receptor (GPCR); N-myristoylation site; Subcellular localization.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

