Structural and functional features of self-assembling protein nanoparticles produced in endotoxin-free Escherichia coli
- PMID: 27059706
- PMCID: PMC4826532
- DOI: 10.1186/s12934-016-0457-z
Structural and functional features of self-assembling protein nanoparticles produced in endotoxin-free Escherichia coli
Abstract
Background: Production of recombinant drugs in process-friendly endotoxin-free bacterial factories targets to a lessened complexity of the purification process combined with minimized biological hazards during product application. The development of nanostructured recombinant materials in innovative nanomedical activities expands such a need beyond plain functional polypeptides to complex protein assemblies. While Escherichia coli has been recently modified for the production of endotoxin-free proteins, no data has been so far recorded regarding how the system performs in the fabrication of smart nanostructured materials.
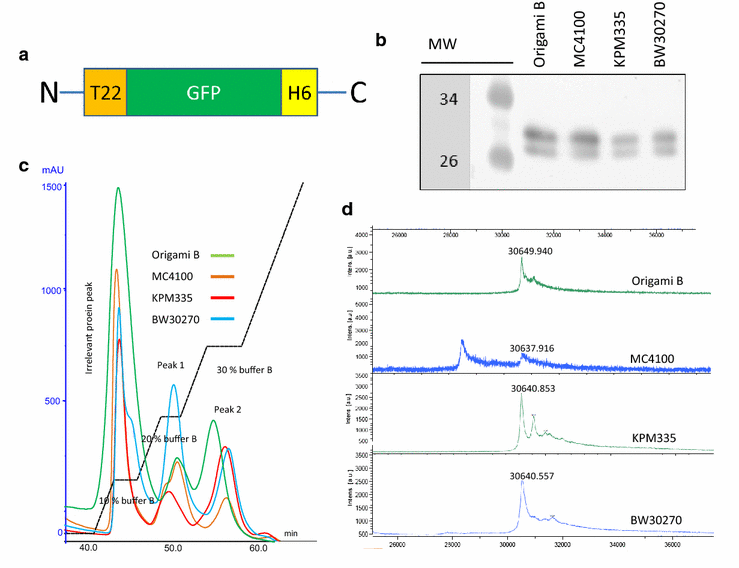
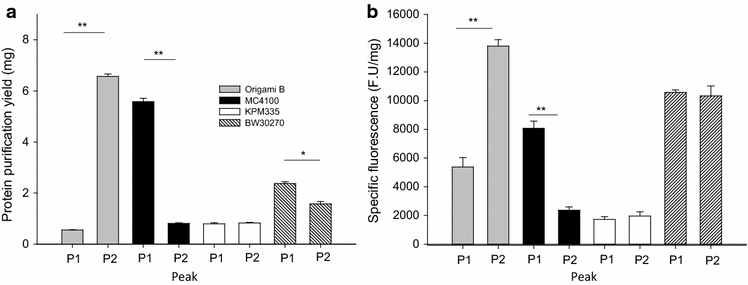
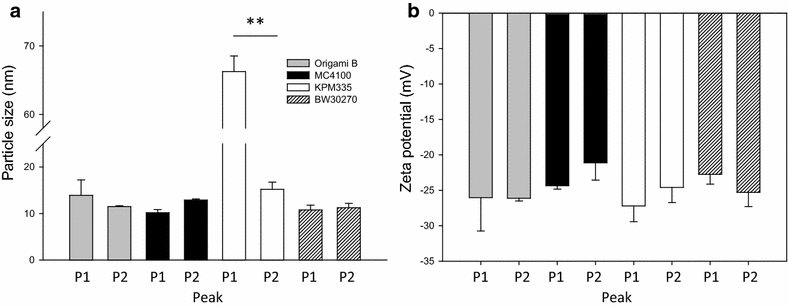
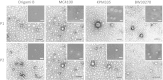
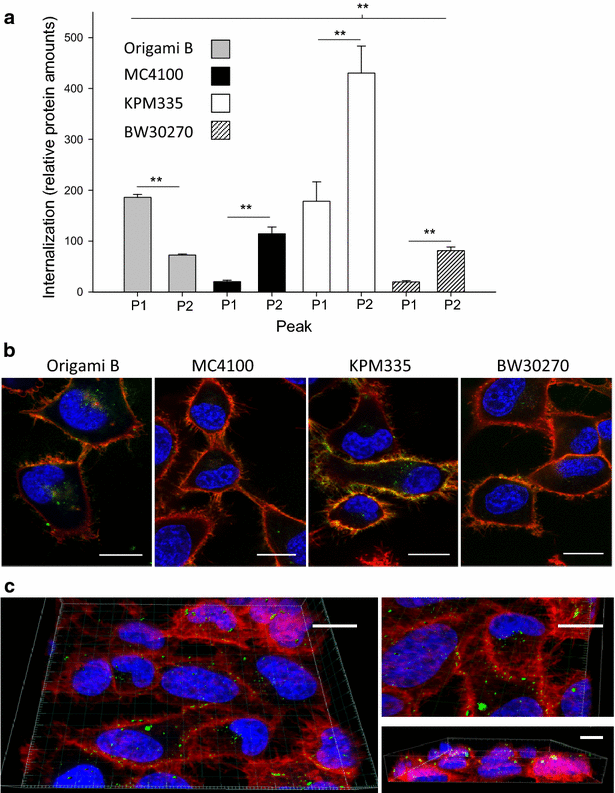
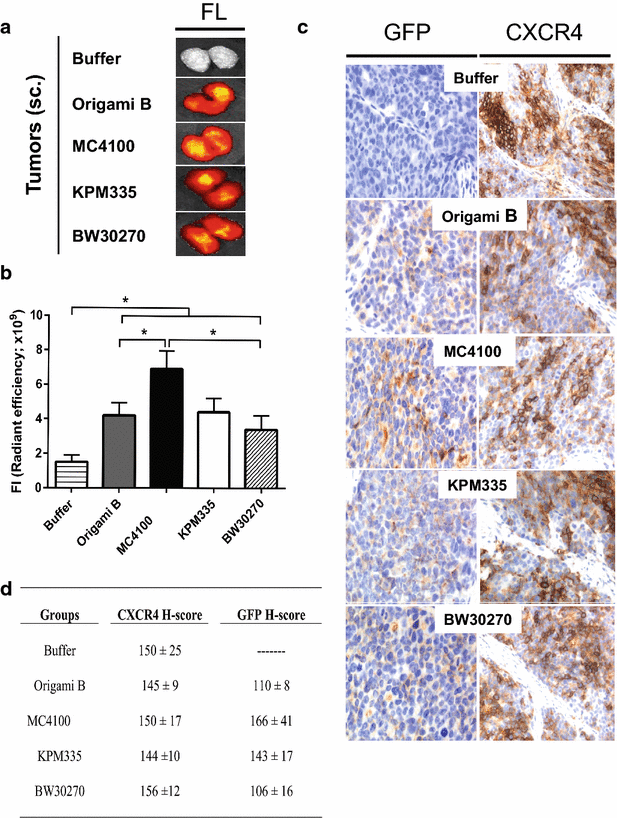
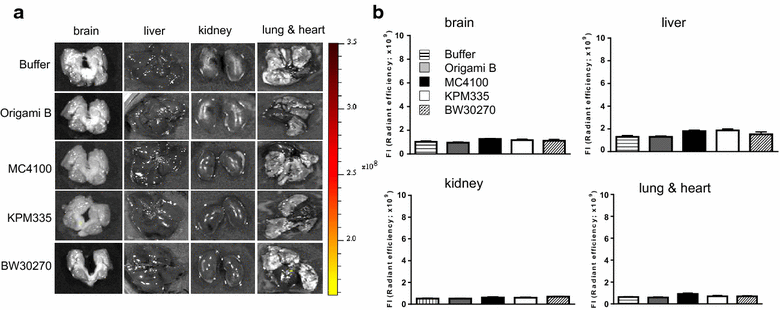
Results: We have here explored the nanoarchitecture and in vitro and in vivo functionalities of CXCR4-targeted, self-assembling protein nanoparticles intended for intracellular delivery of drugs and imaging agents in colorectal cancer. Interestingly, endotoxin-free materials exhibit a distinguishable architecture and altered size and target cell penetrability than counterparts produced in conventional E. coli strains. These variant nanoparticles show an eventual proper biodistribution and highly specific and exclusive accumulation in tumor upon administration in colorectal cancer mice models, indicating a convenient display and function of the tumor homing peptides and high particle stability under physiological conditions.
Discussion: The observations made here support the emerging endotoxin-free E. coli system as a robust protein material producer but are also indicative of a particular conformational status and organization of either building blocks or oligomers. This appears to be promoted by multifactorial stress-inducing conditions upon engineering of the E. coli cell envelope, which impacts on the protein quality control of the cell factory.
Keywords: Biodistribution; Biomaterials; E. coli; Endotoxin-free strains; Nanomedicine; Nanoparticles; Protein engineering; Recombinant proteins.
Figures

Similar articles
-
CXCR4(+)-targeted protein nanoparticles produced in the food-grade bacterium Lactococcus lactis.Nanomedicine (Lond). 2016 Sep;11(18):2387-98. doi: 10.2217/nnm-2016-0200. Epub 2016 Aug 16. Nanomedicine (Lond). 2016. PMID: 27529439
-
In vivo architectonic stability of fully de novo designed protein-only nanoparticles.ACS Nano. 2014 May 27;8(5):4166-76. doi: 10.1021/nn4055732. Epub 2014 Apr 14. ACS Nano. 2014. PMID: 24708510
-
Conformational and functional variants of CD44-targeted protein nanoparticles bio-produced in bacteria.Biofabrication. 2016 Apr 14;8(2):025001. doi: 10.1088/1758-5090/8/2/025001. Biofabrication. 2016. PMID: 27078873
-
Engineering protein self-assembling in protein-based nanomedicines for drug delivery and gene therapy.Crit Rev Biotechnol. 2015 Jun;35(2):209-21. doi: 10.3109/07388551.2013.833163. Epub 2013 Oct 9. Crit Rev Biotechnol. 2015. PMID: 24102113 Review.
-
Membrane Protein Production in E. coli for Applications in Drug Discovery.Adv Exp Med Biol. 2016;896:59-77. doi: 10.1007/978-3-319-27216-0_5. Adv Exp Med Biol. 2016. PMID: 27165319 Review.
Cited by
-
Escherichia coli-derived virus-like particles in vaccine development.NPJ Vaccines. 2017 Feb 9;2:3. doi: 10.1038/s41541-017-0006-8. eCollection 2017. NPJ Vaccines. 2017. PMID: 29263864 Free PMC article.
-
Selective delivery of T22-PE24-H6 to CXCR4+ diffuse large B-cell lymphoma cells leads to wide therapeutic index in a disseminated mouse model.Theranostics. 2020 Apr 6;10(12):5169-5180. doi: 10.7150/thno.43231. eCollection 2020. Theranostics. 2020. PMID: 32373205 Free PMC article.
-
Adenovirus Fibers as Ultra-Stable Vehicles for Intracellular Nanoparticle and Protein Delivery.Biomolecules. 2022 Feb 15;12(2):308. doi: 10.3390/biom12020308. Biomolecules. 2022. PMID: 35204809 Free PMC article.
-
Engineering for an HPV 9-valent vaccine candidate using genomic constitutive over-expression and low lipopolysaccharide levels in Escherichia coli cells.Microb Cell Fact. 2021 Dec 20;20(1):227. doi: 10.1186/s12934-021-01719-8. Microb Cell Fact. 2021. PMID: 34930257 Free PMC article.
-
Functional inclusion bodies produced in the yeast Pichia pastoris.Microb Cell Fact. 2016 Oct 1;15(1):166. doi: 10.1186/s12934-016-0565-9. Microb Cell Fact. 2016. PMID: 27716225 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

