Structural Determinants of Binding the Seven-transmembrane Domain of the Glucagon-like Peptide-1 Receptor (GLP-1R)
- PMID: 27059958
- PMCID: PMC4933217
- DOI: 10.1074/jbc.M116.721977
Structural Determinants of Binding the Seven-transmembrane Domain of the Glucagon-like Peptide-1 Receptor (GLP-1R)
Abstract
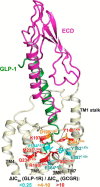
The glucagon-like peptide-1 receptor (GLP-1R) belongs to the secretin-like (class B) family of G protein-coupled receptors. Members of the class B family are distinguished by their large extracellular domain, which works cooperatively with the canonical seven-transmembrane (7TM) helical domain to signal in response to binding of various peptide hormones. We have combined structure-based site-specific mutational studies with molecular dynamics simulations of a full-length model of GLP-1R bound to multiple peptide ligand variants. Despite the high sequence similarity between GLP-1R and its closest structural homologue, the glucagon receptor (GCGR), nearly half of the 62 stably expressed mutants affected GLP-1R in a different manner than the corresponding mutants in GCGR. The molecular dynamics simulations of wild-type and mutant GLP-1R·ligand complexes provided molecular insights into GLP-1R-specific recognition mechanisms for the N terminus of GLP-1 by residues in the 7TM pocket and explained how glucagon-mimicking GLP-1 mutants restored binding affinity for (GCGR-mimicking) GLP-1R mutants. Structural analysis of the simulations suggested that peptide ligand binding mode variations in the 7TM binding pocket are facilitated by movement of the extracellular domain relative to the 7TM bundle. These differences in binding modes may account for the pharmacological differences between GLP-1 peptide variants.
Keywords: G protein-coupled receptor (GPCR); glucagon; molecular dynamics; peptide interaction; site-directed mutagenesis.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Meier J. J. (2012) GLP-1 receptor agonists for individualized treatment of type 2 diabetes mellitus. Nat. Rev. Endocrinol. 8, 728–742 - PubMed
-
- Moller D. E. (2001) New drug targets for type 2 diabetes and the metabolic syndrome. Nature 414, 821–827 - PubMed
-
- Parthier C., Reedtz-Runge S., Rudolph R., and Stubbs M. T. (2009) Passing the baton in class B GPCRs: peptide hormone activation via helix induction? Trends Biochem. Sci. 34, 303–310 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

