A Structurally Novel Chitinase from the Chitin-Degrading Hyperthermophilic Archaeon Thermococcus chitonophagus
- PMID: 27060120
- PMCID: PMC4959179
- DOI: 10.1128/AEM.00319-16
A Structurally Novel Chitinase from the Chitin-Degrading Hyperthermophilic Archaeon Thermococcus chitonophagus
Abstract
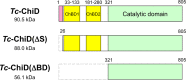
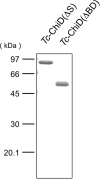
A structurally novel chitinase, Tc-ChiD, was identified from the hyperthermophilic archaeon Thermococcus chitonophagus, which can grow on chitin as the sole organic carbon source. The gene encoding Tc-ChiD contains regions corresponding to a signal sequence, two chitin-binding domains, and a putative catalytic domain. This catalytic domain shows no similarity with previously characterized chitinases but resembles an uncharacterized protein found in the mesophilic anaerobic bacterium Clostridium botulinum Two recombinant Tc-ChiD proteins were produced in Escherichia coli, one without the signal sequence [Tc-ChiD(ΔS)] and the other corresponding only to the putative catalytic domain [Tc-ChiD(ΔBD)]. Enzyme assays using N-acetylglucosamine (GlcNAc) oligomers indicated that both proteins hydrolyze GlcNAc oligomers longer than (GlcNAc)4 Chitinase assays using colloidal chitin suggested that Tc-ChiD is an exo-type chitinase that releases (GlcNAc)2 or (GlcNAc)3 Analysis with GlcNAc oligomers modified with p-nitrophenol suggested that Tc-ChiD recognizes the reducing end of chitin chains. While Tc-ChiD(ΔBD) displayed a higher initial velocity than that of Tc-ChiD(ΔS), we found that the presence of the two chitin-binding domains significantly enhanced the thermostability of the catalytic domain. In T. chitonophagus, another chitinase ortholog that is similar to the Thermococcus kodakarensis chitinase ChiA is present and can degrade chitin from the nonreducing ends. Therefore, the presence of multiple chitinases in T. chitonophagus with different modes of cleavage may contribute to its unique ability to efficiently degrade chitin.
Importance: A structurally novel chitinase, Tc-ChiD, was identified from Thermococcus chitonophagus, a hyperthermophilic archaeon. The protein contains a signal peptide for secretion, two chitin-binding domains, and a catalytic domain that shows no similarity with previously characterized chitinases. Tc-ChiD thus represents a new family of chitinases. Tc-ChiD is an exo-type chitinase that recognizes the reducing end of chitin chains and releases (GlcNAc)2 or (GlcNAc)3 As a thermostable chitinase that recognizes the reducing end of chitin chains was not previously known, Tc-ChiD may be useful in a wide range of enzyme-based technologies to degrade and utilize chitin.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Gooday GW. 1990. Physiology of microbial degradation of chitin and chitosan. Biodegradation 1:177–190. doi:10.1007/BF00058835. - DOI
-
- Bentley SD, Chater KF, Cerdeño-Tárraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147. doi:10.1038/417141a. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

