Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator
- PMID: 27061298
- PMCID: PMC4849733
- DOI: 10.1002/gepi.21965
Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator
Abstract
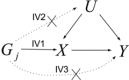
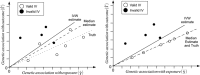
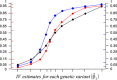
Developments in genome-wide association studies and the increasing availability of summary genetic association data have made application of Mendelian randomization relatively straightforward. However, obtaining reliable results from a Mendelian randomization investigation remains problematic, as the conventional inverse-variance weighted method only gives consistent estimates if all of the genetic variants in the analysis are valid instrumental variables. We present a novel weighted median estimator for combining data on multiple genetic variants into a single causal estimate. This estimator is consistent even when up to 50% of the information comes from invalid instrumental variables. In a simulation analysis, it is shown to have better finite-sample Type 1 error rates than the inverse-variance weighted method, and is complementary to the recently proposed MR-Egger (Mendelian randomization-Egger) regression method. In analyses of the causal effects of low-density lipoprotein cholesterol and high-density lipoprotein cholesterol on coronary artery disease risk, the inverse-variance weighted method suggests a causal effect of both lipid fractions, whereas the weighted median and MR-Egger regression methods suggest a null effect of high-density lipoprotein cholesterol that corresponds with the experimental evidence. Both median-based and MR-Egger regression methods should be considered as sensitivity analyses for Mendelian randomization investigations with multiple genetic variants.
Keywords: Egger regression; Mendelian randomization; instrumental variables; pleiotropy; robust statistics.
© 2016 The Authors. *Genetic Epidemiology Published by Wiley Periodicals, Inc.
Figures

References
-
- Bochud M, Chiolero A, Elston R, Paccaud F. 2008. A cautionary note on the use of Mendelian randomization to infer causation in observational epidemiology. Int J Epidemiol 37(2):414–416. - PubMed
-
- Bowden J, Thompson J, Burton P. 2006. Using pseudo‐data to correct for publication bias in meta‐analysis. Stat Med 25: 3798–3813. - PubMed
-
- Burgess S, Thompson SG. 2015. Mendelian Randomization: Methods for Using Genetic Variants in Causal Estimation. Chapman & Hall, Boca Raton, FL, USA.
-
- Burgess S, Thompson S, CRP CHD Genetics Collaboration . 2011. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol 40(3):755–764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

