RNA-seq transcriptome analysis of a Pseudomonas strain with diversified catalytic properties growth under different culture medium
- PMID: 27061463
- PMCID: PMC4985596
- DOI: 10.1002/mbo3.357
RNA-seq transcriptome analysis of a Pseudomonas strain with diversified catalytic properties growth under different culture medium
Abstract
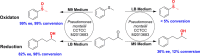
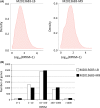
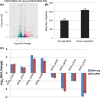
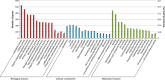
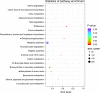
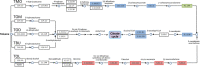
Biocatalysis is an emerging strategy for the production of enantio-pure organic molecules. However, lacking of commercially available enzymes restricts the widespread application of biocatalysis. In this study, we report a Pseudomonas strain which exhibited versatile oxidation activity to synthesize chiral sulfoxides when growing under M9-toluene medium and reduction activity to synthesize chiral alcohols when on Luria-Bertani (LB) medium, respectively. Further comparative transcriptome analysis on samples from these two cultural conditions has identified 1038 differentially expressed genes (DEG). Gene Ontology (GO) enrichment and KEGG pathways analysis demonstrate significant changes in protein synthesis, energy metabolism, and biosynthesis of metabolites when cells cultured under different conditions. We have identified eight candidate enzymes from this bacterial which may have the potential to be used for synthesis of chiral alcohol and sulfoxide chemicals. This work provides insights into the mechanism of diversity in catalytic properties of this Pseudomonas strain growth with different cultural conditions, as well as candidate enzymes for further biocatalysis of enantiomerically pure molecules and pharmaceuticals.
Keywords: Biocatalysis; Pseudomonas; Transcriptome; differentially expressed gene.
© 2016 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures
References
-
- Alcalde, M. 2015. Engineering the ligninolytic enzyme consortium. Trends Biotechnol. 33:155–162. - PubMed
-
- Alcalde, M. , Ferrer M., Plou F. J., and Ballesteros A.. 2006. Environmental biocatalysis: from remediation with enzymes to novel green processes. Trends Biotechnol. 24:281–287. - PubMed
-
- Aldridge, S. 2013. Industry backs biocatalysis for greener manufacturing. Nat. Biotechnol. 31:95–96. - PubMed
-
- Boyd, D. R. , Sharma N. D., McMurray B., Haughey S. A., Allen C. C., Hamilton J. T., et al. 2012. Bacterial dioxygenase‐ and monooxygenase‐catalysed sulfoxidation of benzo[b]thiophenes. Org. Biomol. Chem. 10:782–790. - PubMed
-
- Cao, B. , Nagarajan K., and Loh K. C.. 2009. Biodegradation of aromatic compounds: current status and opportunities for biomolecular approaches. Appl. Microbiol. Biotechnol. 85:207–228. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

