Ligase-4 Deficiency Causes Distinctive Immune Abnormalities in Asymptomatic Individuals
- PMID: 27063650
- PMCID: PMC4842108
- DOI: 10.1007/s10875-016-0266-5
Ligase-4 Deficiency Causes Distinctive Immune Abnormalities in Asymptomatic Individuals
Abstract
Purpose: DNA Ligase 4 (LIG4) is a key factor in the non-homologous end-joining (NHEJ) DNA double-strand break repair pathway needed for V(D)J recombination and the generation of the T cell receptor and immunoglobulin molecules. Defects in LIG4 result in a variable syndrome of growth retardation, pancytopenia, combined immunodeficiency, cellular radiosensitivity, and developmental delay.
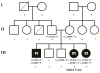
Methods: We diagnosed a patient with LIG4 syndrome by radiosensitivity testing on peripheral blood cells, and established that two of her four healthy siblings carried the same compound heterozygous LIG4 mutations. An extensive analysis of the immune phenotype, cellular radiosensitivity, telomere length, and T and B cell antigen receptor repertoire was performed in all siblings.
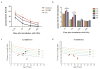
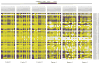
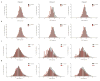
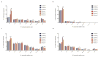
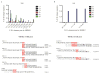
Results: In the three genotypically affected individuals, variable severities of radiosensitivity, alterations of T and B cell counts with an increased percentage of memory cells, and hypogammaglobulinemia, were noticed. Analysis of T and B cell antigen receptor repertoires demonstrated increased usage of alternative microhomology-mediated end-joining (MHMEJ) repair, leading to diminished N nucleotide addition and shorter CDR3 length. However, overall repertoire diversity was preserved.
Conclusions: We demonstrate that LIG4 syndrome presents with high clinical variability even within the same family, and that distinctive immunologic abnormalities may be observed also in yet asymptomatic individuals.
Keywords: DNA repair; Ligase 4 deficiency; immune repertoire; microhomology-mediated end-joining; telomere length.
Conflict of interest statement
None of the authors have any conflicts of interest to declare.
Figures
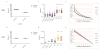
Comment in
-
The Evolving Landscape of Primary Immunodeficiencies.J Clin Immunol. 2016 May;36(4):339-40. doi: 10.1007/s10875-016-0273-6. Epub 2016 Mar 23. J Clin Immunol. 2016. PMID: 27004690 No abstract available.
References
-
- Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA repair. 2004;3(8–9):817–26. - PubMed
-
- Rucci F, Notarangelo LD, Fazeli A, Patrizi L, Hickernell T, Paganini T, et al. Homozygous DNA ligase IV R278H mutation in mice leads to leaky SCID and represents a model for human LIG4 syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(7):3024–9. - PMC - PubMed
-
- Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108(6):781–94. - PubMed
-
- Janeway CA, Jr, TP, Walport M, et al. The generation of diverstity in immunoglobulins. New York: Garland Science; 2001.
-
- Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, et al. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388(6641):492–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

