Inhibition of HSV-1 Replication by Gene Editing Strategy
- PMID: 27064617
- PMCID: PMC4827394
- DOI: 10.1038/srep23146
Inhibition of HSV-1 Replication by Gene Editing Strategy
Abstract
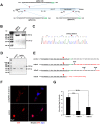
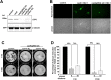
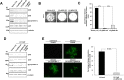
HSV-1 induced illness affects greater than 85% of adults worldwide with no permanent curative therapy. We used RNA-guided CRISPR/Cas9 gene editing to specifically target for deletion of DNA sequences of the HSV-1 genome that span the region directing expression of ICP0, a key viral protein that stimulates HSV-1 gene expression and replication. We found that CRISPR/Cas9 introduced InDel mutations into exon 2 of the ICP0 gene profoundly reduced HSV-1 infectivity in permissive human cell culture models and protected permissive cells against HSV-1 infection. CRISPR/Cas9 mediated targeting ICP0 prevented HSV-1-induced disintegration of promonocytic leukemia (PML) nuclear bodies, an intracellular event critical to productive HSV-1 infection that is initiated by interaction of the ICP0 N-terminus with PML. Combined treatment of cells with CRISPR targeting ICP0 plus the immediate early viral proteins, ICP4 or ICP27, completely abrogated HSV-1 infection. We conclude that RNA-guided CRISPR/Cas9 can be used to develop a novel, specific and efficacious therapeutic and prophylactic platform for targeted viral genomic ablation to treat HSV-1 diseases.
Figures

Similar articles
-
Herpesviral lytic gene functions render the viral genome susceptible to novel editing by CRISPR/Cas9.Elife. 2019 Dec 2;8:e51662. doi: 10.7554/eLife.51662. Elife. 2019. PMID: 31789594 Free PMC article.
-
Cellular Transcriptional Coactivator RanBP10 and Herpes Simplex Virus 1 ICP0 Interact and Synergistically Promote Viral Gene Expression and Replication.J Virol. 2016 Jan 6;90(6):3173-86. doi: 10.1128/JVI.03043-15. J Virol. 2016. PMID: 26739050 Free PMC article.
-
CRISPR/Cas9-Mediated Genome Editing of Herpesviruses Limits Productive and Latent Infections.PLoS Pathog. 2016 Jun 30;12(6):e1005701. doi: 10.1371/journal.ppat.1005701. eCollection 2016 Jun. PLoS Pathog. 2016. PMID: 27362483 Free PMC article.
-
Exploring the potential of genome editing CRISPR-Cas9 technology.Gene. 2017 Jan 30;599:1-18. doi: 10.1016/j.gene.2016.11.008. Epub 2016 Nov 9. Gene. 2017. PMID: 27836667 Review.
-
Development of Genome Editing Approaches against Herpes Simplex Virus Infections.Viruses. 2021 Feb 22;13(2):338. doi: 10.3390/v13020338. Viruses. 2021. PMID: 33671590 Free PMC article. Review.
Cited by
-
Antiviral strategies targeting herpesviruses.J Virus Erad. 2021 May 30;7(3):100047. doi: 10.1016/j.jve.2021.100047. eCollection 2021 Sep. J Virus Erad. 2021. PMID: 34141443 Free PMC article. Review.
-
Therapeutic Application of Genome Editing Technologies in Viral Diseases.Int J Mol Sci. 2022 May 12;23(10):5399. doi: 10.3390/ijms23105399. Int J Mol Sci. 2022. PMID: 35628210 Free PMC article. Review.
-
Therapeutic gene editing: delivery and regulatory perspectives.Acta Pharmacol Sin. 2017 Jun;38(6):738-753. doi: 10.1038/aps.2017.2. Epub 2017 Apr 10. Acta Pharmacol Sin. 2017. PMID: 28392568 Free PMC article. Review.
-
The opportunities and challenges of epigenetic approaches to manage herpes simplex infections.Expert Rev Anti Infect Ther. 2024 Dec;22(12):1123-1142. doi: 10.1080/14787210.2024.2420329. Epub 2024 Nov 6. Expert Rev Anti Infect Ther. 2024. PMID: 39466139 Review.
-
A short overview of CRISPR-Cas technology and its application in viral disease control.Transgenic Res. 2021 Jun;30(3):221-238. doi: 10.1007/s11248-021-00247-w. Epub 2021 Apr 8. Transgenic Res. 2021. PMID: 33830423 Free PMC article. Review.
References
-
- Whitley R. J. & Roizman B. Herpes simplex virus infections. Lancet 357, 1513–1518. - PubMed
-
- Xu F. et al.. Trends in herpes simplex virus type 1 and type 2 seroprevalence in the United States. JAMA 296, 964–973. - PubMed
-
- Baringer J. R. Herpes Virus Encephalitis. In: Davis L. E., Kennedy P. G. E. (eds) Infectious diseases of the nervous system. Butterworth-Heinemann. Oxford pp. 139–164 (2000).
-
- Kennedy P. G. E. & Steiner I. Recent issues in herpes simplex encephalitis. J. Neurovirol. 19, 346–350 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

