Electrostatic and Hydrophobic Interactions Mediate Single-Stranded DNA Recognition and Acta2 Repression by Purine-Rich Element-Binding Protein B
- PMID: 27064749
- PMCID: PMC6368843
- DOI: 10.1021/acs.biochem.6b00006
Electrostatic and Hydrophobic Interactions Mediate Single-Stranded DNA Recognition and Acta2 Repression by Purine-Rich Element-Binding Protein B
Abstract
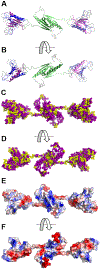
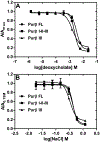
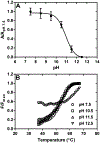
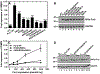
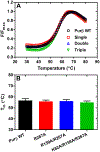
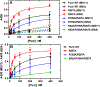
Myofibroblast differentiation is characterized by an increased level of expression of cytoskeletal smooth muscle α-actin. In human and murine fibroblasts, the gene encoding smooth muscle α-actin (Acta2) is tightly regulated by a network of transcription factors that either activate or repress the 5' promoter-enhancer in response to environmental cues signaling tissue repair and remodeling. Purine-rich element-binding protein B (Purβ) suppresses the expression of Acta2 by cooperatively interacting with the sense strand of a 5' polypurine sequence containing an inverted MCAT cis element required for gene activation. In this study, we evaluated the chemical basis of nucleoprotein complex formation between the Purβ repressor and the purine-rich strand of the MCAT element in the mouse Acta2 promoter. Quantitative single-stranded DNA (ssDNA) binding assays conducted in the presence of increasing concentrations of monovalent salt or anionic detergent suggested that the assembly of a high-affinity nucleoprotein complex is driven by a combination of electrostatic and hydrophobic interactions. Consistent with the results of pH titration analysis, site-directed mutagenesis revealed several basic amino acid residues in the intermolecular (R267) and intramolecular (K82 and R159) subdomains that are essential for Purβ transcriptional repressor function in Acta2 promoter-reporter assays. In keeping with their diminished Acta2 repressor activity in fibroblasts, purified Purβ variants containing an R267A mutation exhibited reduced binding affinity for purine-rich ssDNA. Moreover, certain double and triple-point mutants were also defective in binding to the Acta2 corepressor protein, Y-box-binding protein 1. Collectively, these findings establish the repertoire of noncovalent interactions that account for the unique structural and functional properties of Purβ.
Figures

Similar articles
-
Aromatic Residues Dictate the Transcriptional Repressor and Single-Stranded DNA Binding Activities of Purine-Rich Element Binding Protein B.Biochemistry. 2023 Sep 5;62(17):2597-2610. doi: 10.1021/acs.biochem.3c00204. Epub 2023 Aug 9. Biochemistry. 2023. PMID: 37556352 Free PMC article.
-
Purine-rich element binding protein B attenuates the coactivator function of myocardin by a novel molecular mechanism of smooth muscle gene repression.Mol Cell Biochem. 2021 Aug;476(8):2899-2916. doi: 10.1007/s11010-021-04117-1. Epub 2021 Mar 20. Mol Cell Biochem. 2021. PMID: 33743134
-
Structural and functional analysis of single-nucleotide polymorphic variants of purine-rich element-binding protein B.J Cell Biochem. 2019 Apr;120(4):5835-5851. doi: 10.1002/jcb.27869. Epub 2018 Nov 1. J Cell Biochem. 2019. PMID: 30387171 Free PMC article.
-
Structural basis of multisite single-stranded DNA recognition and ACTA2 repression by purine-rich element binding protein B (Purβ).Biochemistry. 2013 Jul 2;52(26):4439-50. doi: 10.1021/bi400283r. Epub 2013 Jun 20. Biochemistry. 2013. PMID: 23724822 Free PMC article.
-
Structure-function analysis of mouse Pur beta II. Conformation altering mutations disrupt single-stranded DNA and protein interactions crucial to smooth muscle alpha-actin gene repression.J Biol Chem. 2007 Dec 7;282(49):35899-909. doi: 10.1074/jbc.M706617200. Epub 2007 Sep 28. J Biol Chem. 2007. PMID: 17906292
Cited by
-
Aromatic Residues Dictate the Transcriptional Repressor and Single-Stranded DNA Binding Activities of Purine-Rich Element Binding Protein B.Biochemistry. 2023 Sep 5;62(17):2597-2610. doi: 10.1021/acs.biochem.3c00204. Epub 2023 Aug 9. Biochemistry. 2023. PMID: 37556352 Free PMC article.
-
Purine-rich element binding protein B attenuates the coactivator function of myocardin by a novel molecular mechanism of smooth muscle gene repression.Mol Cell Biochem. 2021 Aug;476(8):2899-2916. doi: 10.1007/s11010-021-04117-1. Epub 2021 Mar 20. Mol Cell Biochem. 2021. PMID: 33743134
-
Structural and functional analysis of single-nucleotide polymorphic variants of purine-rich element-binding protein B.J Cell Biochem. 2019 Apr;120(4):5835-5851. doi: 10.1002/jcb.27869. Epub 2018 Nov 1. J Cell Biochem. 2019. PMID: 30387171 Free PMC article.
-
Dissection of the interaction between the intrinsically disordered YAP protein and the transcription factor TEAD.Elife. 2017 Apr 21;6:e25068. doi: 10.7554/eLife.25068. Elife. 2017. PMID: 28430104 Free PMC article.
-
Computational investigation of proton transfer, pKa shifts and pH-optimum of protein-DNA and protein-RNA complexes.Proteins. 2017 Feb;85(2):282-295. doi: 10.1002/prot.25221. Epub 2017 Jan 5. Proteins. 2017. PMID: 27936518 Free PMC article.
References
-
- Gabbiani G, Ryan GB, and Majne G (1971) Presence of modified fibroblasts in granulation tissue and their possible role in wound contraction, Experientia 27, 549–550. - PubMed
-
- Hinz B (2007) Formation and function of the myofibroblast during tissue repair, J Invest Dermatol 127, 526–537. - PubMed
-
- Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, and Brown RA (2002) Myofibroblasts and mechano-regulation of connective tissue remodelling, Nat Rev Mol Cell Biol 3, 349–363. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

