How to Predict Molecular Interactions between Species?
- PMID: 27065992
- PMCID: PMC4814556
- DOI: 10.3389/fmicb.2016.00442
How to Predict Molecular Interactions between Species?
Abstract
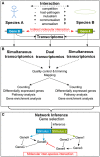
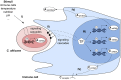
Organisms constantly interact with other species through physical contact which leads to changes on the molecular level, for example the transcriptome. These changes can be monitored for all genes, with the help of high-throughput experiments such as RNA-seq or microarrays. The adaptation of the gene expression to environmental changes within cells is mediated through complex gene regulatory networks. Often, our knowledge of these networks is incomplete. Network inference predicts gene regulatory interactions based on transcriptome data. An emerging application of high-throughput transcriptome studies are dual transcriptomics experiments. Here, the transcriptome of two or more interacting species is measured simultaneously. Based on a dual RNA-seq data set of murine dendritic cells infected with the fungal pathogen Candida albicans, the software tool NetGenerator was applied to predict an inter-species gene regulatory network. To promote further investigations of molecular inter-species interactions, we recently discussed dual RNA-seq experiments for host-pathogen interactions and extended the applied tool NetGenerator (Schulze et al., 2015). The updated version of NetGenerator makes use of measurement variances in the algorithmic procedure and accepts gene expression time series data with missing values. Additionally, we tested multiple modeling scenarios regarding the stimuli functions of the gene regulatory network. Here, we summarize the work by Schulze et al. (2015) and put it into a broader context. We review various studies making use of the dual transcriptomics approach to investigate the molecular basis of interacting species. Besides the application to host-pathogen interactions, dual transcriptomics data are also utilized to study mutualistic and commensalistic interactions. Furthermore, we give a short introduction into additional approaches for the prediction of gene regulatory networks and discuss their application to dual transcriptomics data. We conclude that the application of network inference on dual-transcriptomics data is a promising approach to predict molecular inter-species interactions.
Keywords: dual RNA-seq; dual transcriptomics; gene regulatory network; host-pathogen interaction; molecular inter-species interaction; network inference.
Figures
Similar articles
-
Computational prediction of molecular pathogen-host interactions based on dual transcriptome data.Front Microbiol. 2015 Feb 6;6:65. doi: 10.3389/fmicb.2015.00065. eCollection 2015. Front Microbiol. 2015. PMID: 25705211 Free PMC article.
-
Data- and knowledge-based modeling of gene regulatory networks: an update.EXCLI J. 2015 Mar 2;14:346-78. doi: 10.17179/excli2015-168. eCollection 2015. EXCLI J. 2015. PMID: 27047314 Free PMC article. Review.
-
An Interspecies Regulatory Network Inferred from Simultaneous RNA-seq of Candida albicans Invading Innate Immune Cells.Front Microbiol. 2012 Mar 12;3:85. doi: 10.3389/fmicb.2012.00085. eCollection 2012. Front Microbiol. 2012. PMID: 22416242 Free PMC article.
-
Two's company: studying interspecies relationships with dual RNA-seq.Curr Opin Microbiol. 2018 Apr;42:7-12. doi: 10.1016/j.mib.2017.09.001. Epub 2017 Sep 26. Curr Opin Microbiol. 2018. PMID: 28957710 Review.
-
dRNASb: a systems biology approach to decipher dynamics of host-pathogen interactions using temporal dual RNA-seq data.Microb Genom. 2022 Sep;8(9):mgen000862. doi: 10.1099/mgen.0.000862. Microb Genom. 2022. PMID: 36136078 Free PMC article.
Cited by
-
Humic-acid-driven escape from eye parasites revealed by RNA-seq and target-specific metabarcoding.Parasit Vectors. 2020 Aug 28;13(1):433. doi: 10.1186/s13071-020-04306-9. Parasit Vectors. 2020. PMID: 32859251 Free PMC article.
-
Integrating Large-Scale Data and RNA Technology to Protect Crops from Fungal Pathogens.Front Plant Sci. 2016 May 31;7:631. doi: 10.3389/fpls.2016.00631. eCollection 2016. Front Plant Sci. 2016. PMID: 27303409 Free PMC article. Review.
-
Mathematical models to study the biology of pathogens and the infectious diseases they cause.iScience. 2022 Mar 15;25(4):104079. doi: 10.1016/j.isci.2022.104079. eCollection 2022 Apr 15. iScience. 2022. PMID: 35359802 Free PMC article. Review.
-
Resolving host-pathogen interactions by dual RNA-seq.PLoS Pathog. 2017 Feb 16;13(2):e1006033. doi: 10.1371/journal.ppat.1006033. eCollection 2017 Feb. PLoS Pathog. 2017. PMID: 28207848 Free PMC article. Review.
-
Charting Peptide Shared Sequences Between 'Diabetes-Viruses' and Human Pancreatic Proteins, Their Structural and Autoimmune Implications.Bioinform Biol Insights. 2024 Nov 5;18:11779322241289936. doi: 10.1177/11779322241289936. eCollection 2024. Bioinform Biol Insights. 2024. PMID: 39502449 Free PMC article.
References
-
- Andrews S. (2010). Fastqc: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
-
- Asai S., Rallapalli G., Piquerez S. J. M., Caillaud M.-C., Furzer O. J., Ishaque N., et al. . (2014). Expression profiling during arabidopsis/downy mildew interaction reveals a highly-expressed effector that attenuates responses to salicylic acid. PLoS Pathog. 10:e1004443. 10.1371/journal.ppat.1004443 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

