To Be Specific or Not: The Critical Relationship Between Hox And TALE Proteins
- PMID: 27066866
- PMCID: PMC4875764
- DOI: 10.1016/j.tig.2016.03.004
To Be Specific or Not: The Critical Relationship Between Hox And TALE Proteins
Abstract
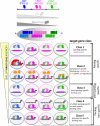
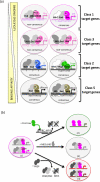
Hox proteins are key regulatory transcription factors that act in different tissues of the embryo to provide specific spatial and temporal coordinates to each cell. These patterning functions often depend on the presence of the TALE-homeodomain class cofactors, which form cooperative DNA-binding complexes with all Hox proteins. How this family of cofactors contributes to the highly diverse and specific functions of Hox proteins in vivo remains an important unsolved question. We review here the most recent advances in understanding the molecular mechanisms underlying Hox-TALE function. In particular, we discuss the role of DNA shape, DNA-binding affinity, and protein-protein interaction flexibility in dictating Hox-TALE specificity. We propose several models to explain how these mechanisms are integrated with each other in the context of the many distinct functions that Hox and TALE factors carry out in vivo.
Keywords: DNA-binding specificity; Hox proteins; SELEX-seq; SPIMs; TALE cofactors; homeodomains.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures


References
-
- Pick L, Heffer A. Hox gene evolution: Multiple mechanisms contributing to evolutionary novelties. Ann. N. Y. Acad. Sci. 2012;1256:15–32. - PubMed
-
- Hombría JC-G, Lovegrove B. Beyond homeosis--HOX function in morphogenesis and organogenesis. Differentiation. 2003;71:461–76. - PubMed
-
- Holland PWH. Evolution of homeobox genes. Wiley Interdisciplinary Reviews: Developmental Biology. 2013;2:31–45. - PubMed
-
- Merabet S, et al. Classification of sequence signatures: a guide to Hox protein function. Bioessays. 2009;31:500–511. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

