A miR-372/let-7 Axis Regulates Human Germ Versus Somatic Cell Fates
- PMID: 27066911
- PMCID: PMC5145290
- DOI: 10.1002/stem.2378
A miR-372/let-7 Axis Regulates Human Germ Versus Somatic Cell Fates
Abstract
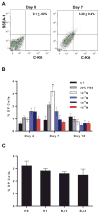
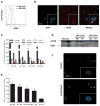
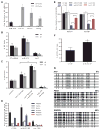
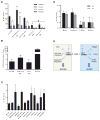
The embryonic stem cell cycle (ESCC) and let-7 families of miRNAs function antagonistically in the switch between mouse embryonic stem cell self-renewal and somatic differentiation. Here, we report that the human ESCC miRNA miR-372 and let-7 act antagonistically in germline differentiation from human embryonic stem cells (hESCs) and human induced pluripotent stem cells (iPSCs). hESC and iPSC-derived primordial germ cell-like cells (PGCLCs) expressed high levels of miR-372 and conversely, somatic cells expressed high levels of let-7. Manipulation of miRNA levels by introduction of miRNA mimics or knockdown with miRNA sponges demonstrated that miR-372 promotes whereas let-7 antagonizes PGCLC differentiation. Knockdown of the individual miR-372 targets SMARCC1, MECP2, CDKN1, RBL2, RHOC, and TGFBR2 increased PGCLC production, whereas knockdown of the let-7 targets CMYC and NMYC suppressed PGCLC differentiation. These findings uncover a miR-372/let-7 axis regulating human primordial germ cell (PGC) specification. Stem Cells 2016;34:1985-1991.
Keywords: Embryonic stem cells; Primordial germ cells; let-7; microRNA; mir-372.
© 2016 AlphaMed Press.
Figures
Similar articles
-
Exogenous supplementation of Activin A enhances germ cell differentiation of human embryonic stem cells.Mol Hum Reprod. 2015 May;21(5):410-23. doi: 10.1093/molehr/gav004. Epub 2015 Jan 29. Mol Hum Reprod. 2015. PMID: 25634576
-
Embryonic stem cell-related miRNAs are involved in differentiation of pluripotent cells originating from the germ line.Mol Hum Reprod. 2010 Nov;16(11):793-803. doi: 10.1093/molehr/gaq053. Epub 2010 Jun 21. Mol Hum Reprod. 2010. PMID: 20566704
-
Opposing microRNA families regulate self-renewal in mouse embryonic stem cells.Nature. 2010 Feb 4;463(7281):621-6. doi: 10.1038/nature08725. Epub 2010 Jan 6. Nature. 2010. PMID: 20054295 Free PMC article.
-
Germline specification from pluripotent stem cells.Stem Cell Res Ther. 2022 Feb 21;13(1):74. doi: 10.1186/s13287-022-02750-1. Stem Cell Res Ther. 2022. PMID: 35189957 Free PMC article. Review.
-
Using Embryo Models to Understand the Development and Progression of Embryonic Lineages: A Focus on Primordial Germ Cell Development.Cells Tissues Organs. 2024;213(6):503-522. doi: 10.1159/000538275. Epub 2024 Mar 13. Cells Tissues Organs. 2024. PMID: 38479364 Free PMC article. Review.
Cited by
-
Mutual Regulation of ncRNAs and Chromatin Remodeling Complexes in Normal and Pathological Conditions.Int J Mol Sci. 2023 Apr 25;24(9):7848. doi: 10.3390/ijms24097848. Int J Mol Sci. 2023. PMID: 37175555 Free PMC article. Review.
-
MiRNA extraction from cell-free biofluid using protein corona formed around carboxyl magnetic nanoparticles.ACS Biomater Sci Eng. 2018 Feb 12;4(2):654-662. doi: 10.1021/acsbiomaterials.7b00668. Epub 2017 Dec 18. ACS Biomater Sci Eng. 2018. PMID: 29623292 Free PMC article.
-
MiR-202-5p is a novel germ plasm-specific microRNA in zebrafish.Sci Rep. 2017 Aug 1;7(1):7055. doi: 10.1038/s41598-017-07675-x. Sci Rep. 2017. PMID: 28765643 Free PMC article.
-
Small RNA in situ hybridization in Caenorhabditis elegans, combined with RNA-seq, identifies germline-enriched microRNAs.Dev Biol. 2016 Oct 15;418(2):248-257. doi: 10.1016/j.ydbio.2016.08.003. Epub 2016 Aug 10. Dev Biol. 2016. PMID: 27521456 Free PMC article.
-
The Zinc Finger Transcription Factor PLAGL2 Enhances Stem Cell Fate and Activates Expression of ASCL2 in Intestinal Epithelial Cells.Stem Cell Reports. 2018 Aug 14;11(2):410-424. doi: 10.1016/j.stemcr.2018.06.009. Epub 2018 Jul 12. Stem Cell Reports. 2018. PMID: 30017821 Free PMC article.
References
-
- Geijsen N, Horoschak M, Kim K, et al. Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature. 2004;427:148–154. - PubMed
-
- Sasaki K, Yokobayashi S, Nakamura T, et al. Robust In Vitro Induction of Human Germ Cell Fate from Pluripotent Stem Cells. Cell Stem Cell. 2015;17:178–194. - PubMed
SUPPLEMENTARY REFERENCES
-
- West FD, Machacek DW, Boyd NL, et al. Enrichment and differentiation of human germ-like cells mediated by feeder cells and basic fibroblast growth factor signaling. Stem Cells. 2008;26:2768–2776. - PubMed
-
- Judson H, Hayward BE, Sheridan E, et al. A global disorder of imprinting in the human female germ line. Nature. 2002;416:539–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

