Transcription factors in microalgae: genome-wide prediction and comparative analysis
- PMID: 27067009
- PMCID: PMC4827209
- DOI: 10.1186/s12864-016-2610-9
Transcription factors in microalgae: genome-wide prediction and comparative analysis
Abstract
Background: Studying transcription factors, which are some of the key players in gene expression, is of outstanding interest for the investigation of the evolutionary history of organisms through lineage-specific features. In this study we performed the first genome-wide TF identification and comparison between haptophytes and other algal lineages.
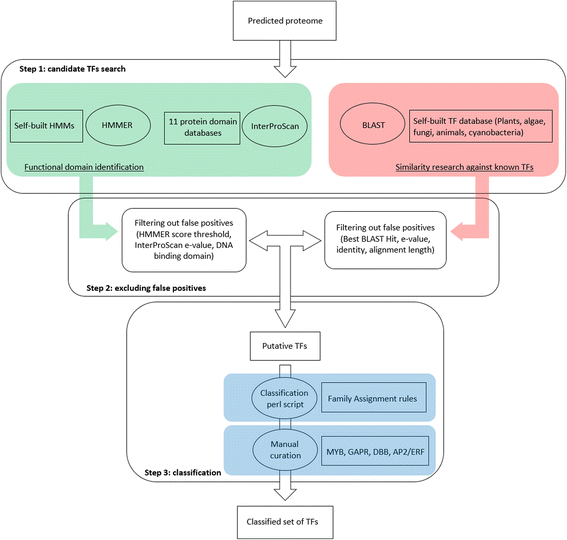
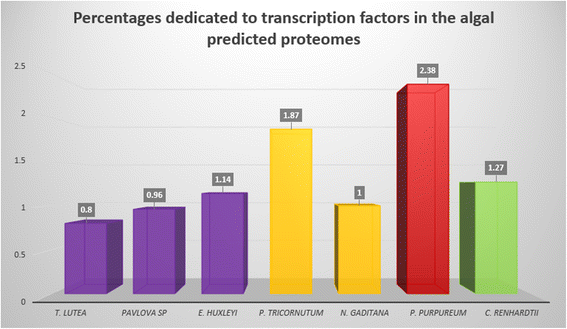
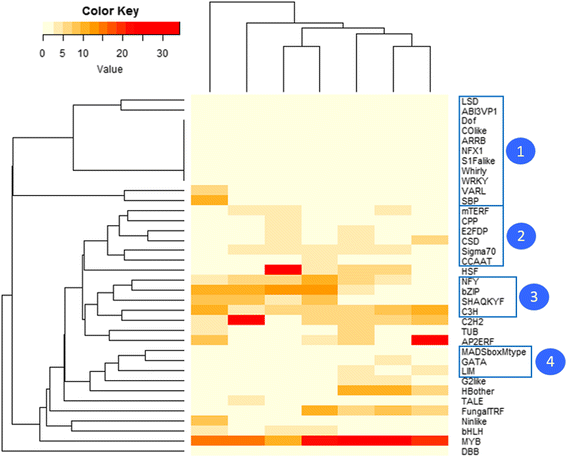
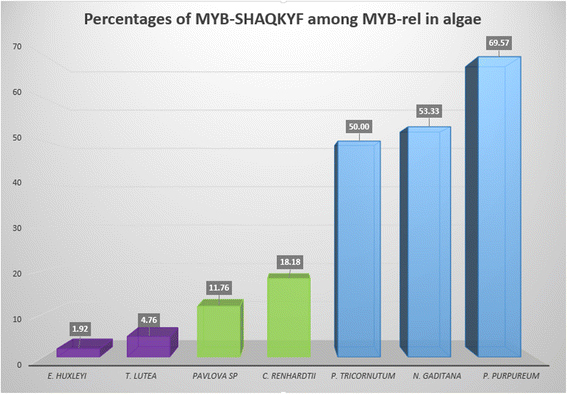
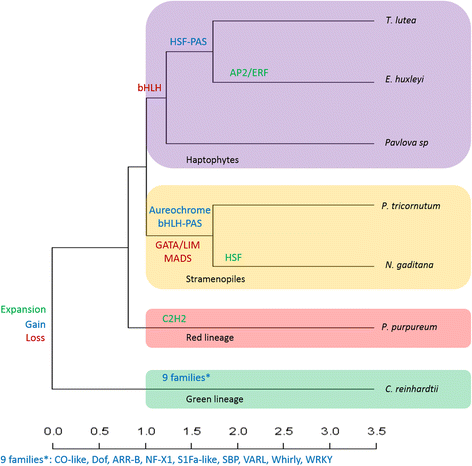
Results: For TF identification and classification, we created a comprehensive pipeline using a combination of BLAST, HMMER and InterProScan software. The accuracy evaluation of the pipeline shows its applicability for every alga, plant and cyanobacterium, with very good PPV and sensitivity. This pipeline allowed us to identify and classified the transcription factor complement of the three haptophytes Tisochrysis lutea, Emiliania huxleyi and Pavlova sp.; the two stramenopiles Phaeodactylum tricornutum and Nannochloropsis gaditana; the chlorophyte Chlamydomonas reinhardtii and the rhodophyte Porphyridium purpureum. By using T. lutea and Porphyridium purpureum, this work extends the variety of species included in such comparative studies, allowing the detection and detailed study of lineage-specific features, such as the presence of TF families specific to the green lineage in Porphyridium purpureum, haptophytes and stramenopiles. Our comprehensive pipeline also allowed us to identify fungal and cyanobacterial TF families in the algal nuclear genomes.
Conclusions: This study provides examples illustrating the complex evolutionary history of algae, some of which support the involvement of a green alga in haptophyte and stramenopile evolution.
Keywords: Algae; Endosymbiotic gene transfer; Haptophytes; Prediction pipeline; Stramenopiles; Tisochrysis lutea; Transcription factors.
Figures

References
-
- Heydarizadeh P, Marchand J, Chenais B, Sabzalian MR, Zahedi M, Moreau B, Schoefs B. Functional investigations in diatoms need more than a transcriptomic approach. Diatom Res. 2014;29:75–89. doi: 10.1080/0269249X.2014.883727. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

